CRIB Configuration
Artemis configuration varies from experiment to experiment. We would like to explain in this chapter how they are configured and used in CRIB experiment.
Artemis configuration varies from experiment to experiment. We would like to explain in this chapter how they are configured and used in CRIB experiment.
CRIB shares the analysis environment of all experiments under one user account (username crib). Therefore, when you want to check data from an old experiment or when several people are analysing the data, you need to log in to the same user account.
Of course, the analysis environment varies according to the experiment (and even different environments for different users within the same experiment!) and these have to be managed well. The “.bashrc/.zshrc” and “artlogin (artlogin2)” commands set them up. Currently we are using “zsh (.zshrc)”.
export EXP_NAME="current" # your experiment
export EXP_NAME_OLD="previous" # old experimentThe EXP_NAME is current experiment and you can enter the environment by using artlogin command.
At the same time, the EXP_NAME_OLD is the old experiment and you can use artlogin2 command.
In the current version, we support two experimental environment and if you want to check other experimental data, please change EXP_NAME_OLD.
When you modify “.bashrc/.zshrc”, all people’s settings will change.
Therefore please do not change EXP_NAME as much as possible, because we want to set this environment variable as the active experiment.
If you change this, please report it so that CRIB members are aware of it.
Commands may be created in the future to enter the environment of all experiments flexibly, not just two. (like artoldlogin {expname} {username}?)
Then you can enter the different analysis environment like this:
> artlogin (username)
> artlogin2 (username)CRIB uses a default user as well as individual analysis environments. The username of the default user is the same with experiment name.
If you set the name of the experiment to “si26a” (EXP_NAME), then the username “si26a” will be the default user.
The user’s environment can be entered with the “artlogin” command with no arguments.
> artlogin
> pwd
/home/crib/art_analysis/si26a/si26aIf you want to test something by changing files, or if you want to use your own VNC server, you can enter that environment by specifying its name as an argument.
> artlogin okawa # if this is the first time to command, you will see setup comments.
> pwd
/home/crib/art_analysis/si26a/okawaWhen using the default user, try to avoid using a VNC server (do not create .vncdisplay files). The main reason for creating a default user is to analyse locally (for shifters) in the online analysis, and using a VNC server makes it impossible to view the figures locally.
The directory structure comprising artemis is as follows. (The location of artemis itself is omitted).
> tree -L 2 ~/art_analysis
/home/crib/art_analysis
├── current # accessed by "artlogin"
│ ├── current # default user
│ └── okawa # individual user
├── previous # accessed by "artlogin2"
│ ├── previous
│ └── okawa
├── old1
│ ├── old1
│ └── okawa
└── old2
# -- snip --We often use “nssta” (non-save mode start) analysis in the beam tuning.
It is not necessary to take data, but we need to check the beam condition by using artemis.
In this case, TRIDFEventStore can be used as online mode.
By default, if we don’t add an input file name and set the SHMID (Shared Memory ID), artemis will use online mode. However, it is necessary to use different types of steering files, one for use in online-mode and the other for use from a file, which can be complicated…
Therefore, the same steering file was changed to automatically go online mode when the ridf file was not present.
# from ridf files
artemis [0] add steering/hoge.yaml NAME=hoge NUM=0000# online-mode
artemis [0] add steering/hoge.yaml # no argumentTo achieve this, the original file was changed as follows.
129 for (Int_t i=0; i!=n;i++) {
130 printf("file = %s\n",fFileName[i].Data());
131+ if(!gSystem->FindFile(".", fFileName[i])) {
132+ Info("Init", "No input file -> Online mode");
133+ fIsOnline = kTRUE;
134+ }
135 }
We always use SHMID=0, so it works simply by adding the following sentence.
- name: ridf
type: art::TRIDFEventStore
parameter:
OutputTransparency: 1
InputFiles:
- *input
SHMID: 0still under consideration in this part!
CRIB often wants to customise artemis because it originally used ANAPAW and wants to perform analysis like ANAPAW. However, we do not want to make too many changes to the source code of artemis itself, and we want to make it work in the user-defined part. (it means in the artemis work directory)
In particular, it is often the case that we want to create a new artemis command, but writing the command source on the work directory and registering it in artemislogon.C did not work somehow…
Also, artemislogon.C is automatically generated (from .artemislogon.C.in) by the cmake functionality, and even if this itself is changed, it will revert when cmake is redone.
Therefore, a file called userlogon.C was prepared, which only took out the user-defined part from artemislogon.C.
The following files have been modified to read this.
14 #include <TInterpreter.h>
15+#include <TSystem.h>
16 #include "TLoopManager.h"
44 TRint::ProcessLine(".x artemislogon.C");
45+ FileStat_t info;
46+ if (gSystem->GetPathInfo("userlogon.C", info)==0) {
47+ TRint::ProcessLine(".x userlogon.C");
48+ }
If there is a userlogon.C file in the work directory, it is loaded, otherwise artemis can be used as usual.
This file can be used freely! What we wanted to do most is to register user-defined commands, which can be done as follows.
{
// load user function
gROOT->ProcessLine(".L macro/UserUtil.C");
// User commands register
// cf definition: TCatCmdFactory *cf = TCatCmdFactory::Instance();
cf->Register(TCatCmdLoopStart::Instance());
cf->Register(TCatCmdLoopStop::Instance());
cf->Register(new art::TCmdXfitg);
cf->Register(new art::TCmdXstatus);
cf->Register(new art::TCmdXYblow);
cf->Register(new art::TCmdXblow);
cf->Register(TCatCmdTCutG::Instance());
cf->Register(new art::TCmdErase);
cf->Register(new art::TCmdDraw);
// TTree merge setting
TTree::SetMaxTreeSize( 1000000000000LL ); // 1TB
}The first line gROOT->ProcessLine(".L macro/UserUtil.C") load the user definition function.
You can add any function to the “macro/UserUtil.C” file, and the function to load TCutG object in “/gate/*.root” directory is written defaultly.
For more detail, please see tcutg command and gate pages.
(For some reason, an error occurred when writing in artemislogon.C.) You can also customise it in other ways to make it easier for you. For example, when creating a TTree, a setting to increase the file size limit is also included by default.
Various commands (mainly the same with ANAPAW commands) have been developed for CRIB experiment. For more information, please click here (src-crib/commands). These commands are registered in userlogon.C. (See previous section.)
This section explains how to use them.
the default figures:

This is exactly the same as the resume command, because ANAPAW starts the event loop with start instead of resume.
This is exactly the same as the suspend command, because ANAPAW stops the event loop with stop instead of suspend.
For 1D histograms, by selecting the two ends of two points, the peak between them is fitted with a Gaussian.
artemis [7] xf
Info in <art::TCmdXfitg::Cmd>: click on the lowest edge:
Info in <art::TCmdXfitg::Cmd>: click on the highest edge:
Info in <art::TCmdXfitg::Cmd>: X1: -1437.56, X2: -1419.11
FCN=81.6642 FROM MIGRAD STATUS=CONVERGED 71 CALLS 72 TOTAL
EDM=3.35095e-09 STRATEGY= 1 ERROR MATRIX ACCURATE
EXT PARAMETER STEP FIRST
NO. NAME VALUE ERROR SIZE DERIVATIVE
1 Constant 1.16439e+03 2.43862e+01 8.08454e-02 9.04256e-07
2 Mean -1.43081e+03 4.54001e-02 6.82262e-04 -1.74034e-03
3 Sigma 2.81435e+00 4.07888e-02 1.55351e-05 -3.15946e-03
artemis [8]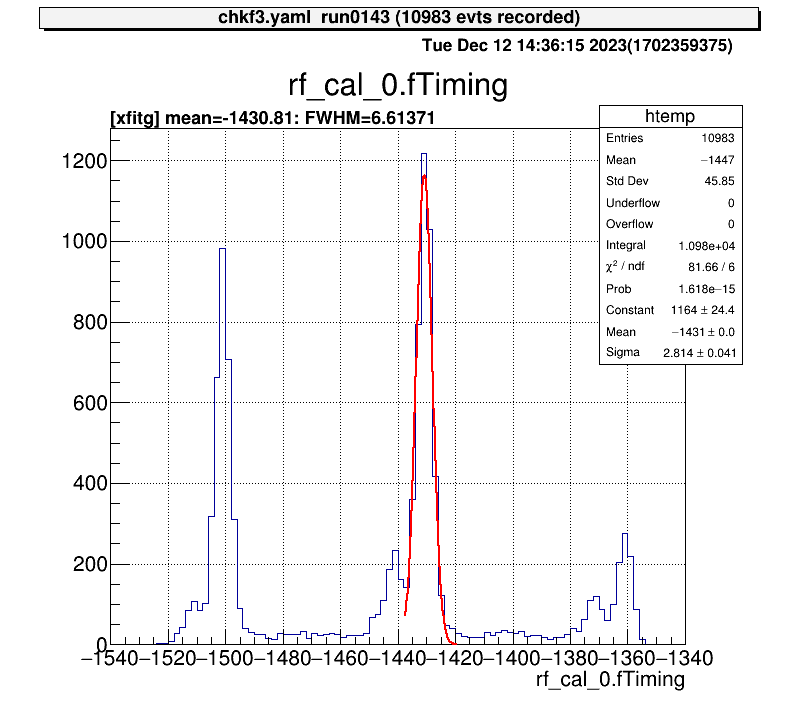
For 1D histograms, select both ends and crop the histogram between them.
artemis [10] xblo
Info in <art::TCmdXblow::Run>: click on the lowest edge:
Info in <art::TCmdXblow::Run>: click on the highest edge:
Info in <art::TCmdXblow::Run>: X1: -1439.3, X2: -1417.37
Info in <art::TCmdXblow::Run>: id = 2 hist is created
artemis [11]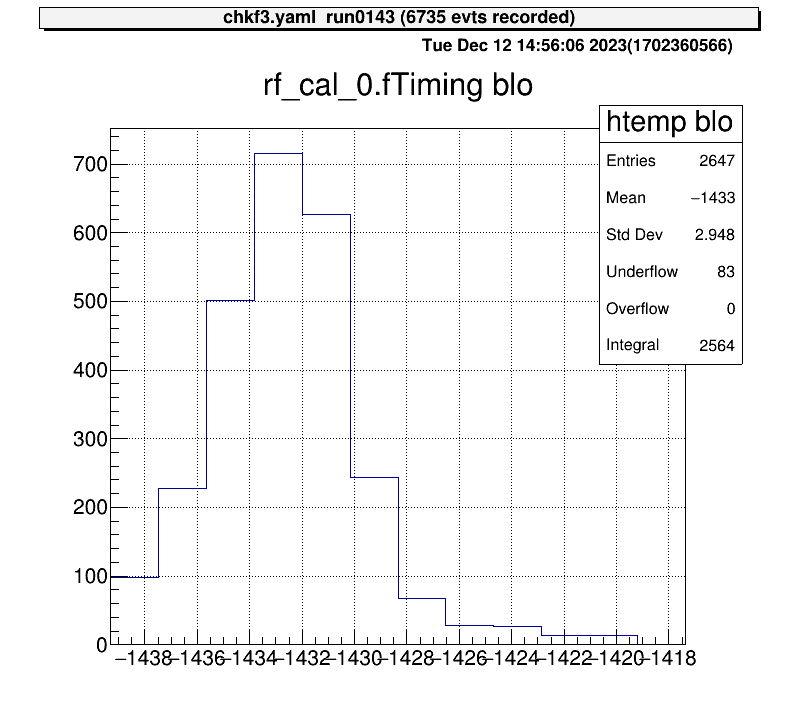
For 2D histograms, select both corners and crop the histogram between them.
artemis [60] xyblo
Info in <art::TCmdXYblow::Run>: click on one corner:
Info in <art::TCmdXYblow::Run>: X1: 9.2154, Y1: 46.6159
Info in <art::TCmdXYblow::Run>: click on the other corner:
Info in <art::TCmdXYblow::Run>: X2: 21.7032, Y2: 23.952
Info in <art::TCmdXYblow::Run>: id = 6 hist is created
artemis [61]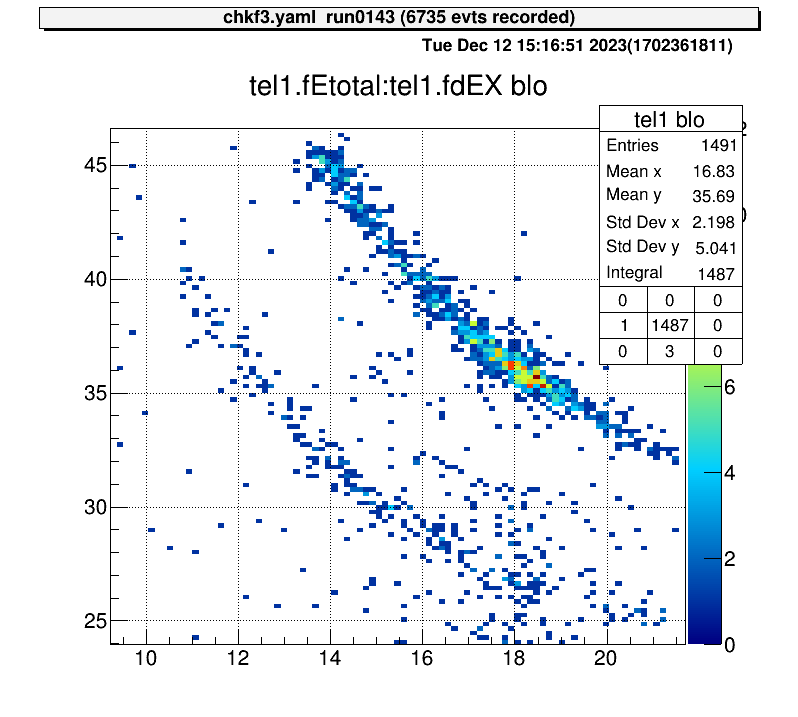
For 2D histograms, select both corners and determine the ratio of the total number of events.
artemis [8] xs
Info in <art::TCmdXstatus::Cmd>: click on one corner:
Info in <art::TCmdXstatus::Cmd>: X1: 14.1496, Y1: 41.4826
Info in <art::TCmdXstatus::Cmd>: click on the other corner:
Info in <art::TCmdXstatus::Cmd>: X2: 21.0941, Y2: 31.9909
------------------
selected = 976, total = 7526
ratio = 0.129684 (12.9684%)
artemis [9]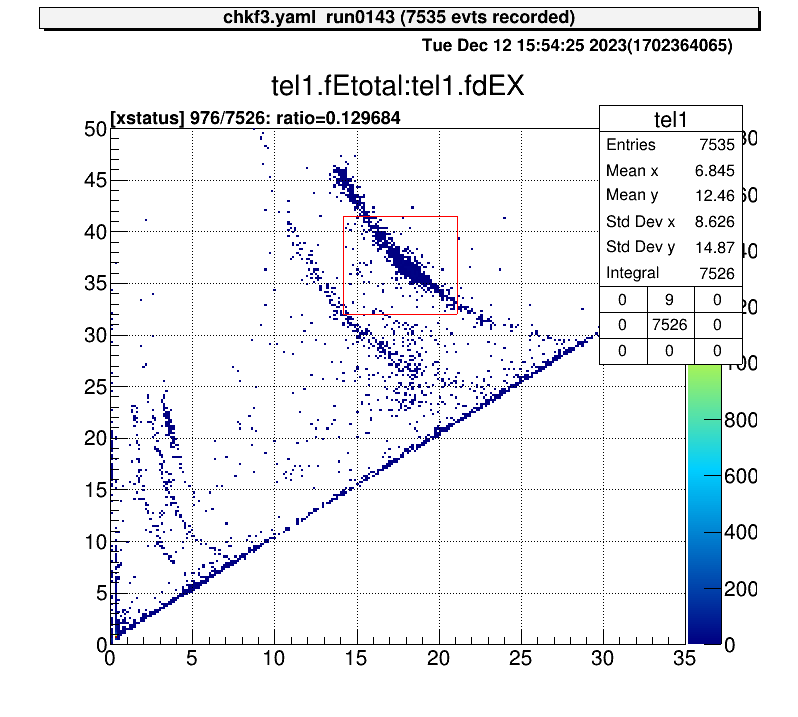
For 2D histograms, this command create TCutG object and store in a ROOT file.
If you select to save the object, the file will place to the gate/*.root directory.
There objects are automatically loaded. (please check user config page.)
This is the example how to use this command.
artemis [] ht something
artemis [] tc
Info in <TCatCmdTCutG::Cmd>: Xaxis name : f2ppac.fX Yaxis name : f2ppac.fY
Info in <TCatCmdTCutG::Cmd>: When you have finished specifying the area (last point), double-click on it.
Info in <TCatCmdTCutG::Cmd>: (x, y) = (9.050404, 10.301410)
Info in <TCatCmdTCutG::Cmd>: (x, y) = (5.047341, -8.294592)
Info in <TCatCmdTCutG::Cmd>: (x, y) = (-12.183236, -3.839300)
Info in <TCatCmdTCutG::Cmd>: (x, y) = (3.306878, -15.074384)
Info in <TCatCmdTCutG::Cmd>: (x, y) = (-3.306878, -32.120720)
Info in <TCatCmdTCutG::Cmd>: (x, y) = (9.920635, -15.461801)
Info in <TCatCmdTCutG::Cmd>: (x, y) = (18.274854, -29.989928)
Info in <TCatCmdTCutG::Cmd>: (x, y) = (16.186299, -11.200217)
Info in <TCatCmdTCutG::Cmd>: (x, y) = (35.157338, -4.420425)
Info in <TCatCmdTCutG::Cmd>: (x, y) = (14.271791, -4.807841)
Info in <TCatCmdTCutG::Cmd>: (x, y) = (10.964912, 9.332869)
Info in <TCatCmdTCutG::Cmd>: (x, y) = (10.964912, 9.332869)
if you want to save it, input the TCutG name [name/exit] f2star
Info in <TCatCmdTCutG::Cmd>: Created gate/f2star.root
To select an area, click on the vertices of the area you want to select, then double-click at the last vertex.
If you want to save this object, enter the “cut” name.
In this example, I input the f2star as the object name.
If you don’t want to save, enter “exit”.
Then the gate/f2star.root will be created.
And after reload the artemis, the gate will be loaded automatically and we can use histogram definition and “tree->Draw” selection part.
For the detail please check gate page.
artemis [] tree->Draw("f2ppac.fY:f2ppac.fX>>(200,-50.,50., 200,-50.,50.)","f2star","colz")
Grammar issue I think.
export LD_LIBRARY_PATH=$TARTSYS/lib:$LD_LIBRARY_PATH
-if [ "@BUILD_GET@" == "ON" ]; then
+if [[ "@BUILD_GET@" == "ON" ]]; then
export LD_LIBRARY_PATH=@GET_LIB_DIR@:$LD_LIBRARY_PATH
fi
-if [ "@MPI_CXX_FOUND@" == "TRUE" ]; then
+if [[ "@MPI_CXX_FOUND@" == "TRUE" ]]; then
dir=@MPI_CXX_LIBRARIES@
libdir="$(dirname $dir)"
Add cross hair.
84 void TCatCmdXval::GetEvent()
85 {
86+ dynamic_cast<TPad *>(gPad)->DrawCrosshair();
87 const int event = gPad->GetEvent();
After the command, the projected histogram will automatically be displayed.
55 if (!obj->InheritsFrom(TH2::Class())) {
56 // TArtCore::Info("TCatCmdPr::Run","%s is not 2D histogram",
57 // obj->GetName());
58+ Info("Run", "%s is not 2D histogram", obj->GetName());
59 continue;
60 }
61+ Int_t nid = (gDirectory->GetList())->GetEntries();
62 Run((TH2*) obj, opt);
63+ Info("Run", "id = %d hist is created", nid);
64+ TCatHistManager::Instance()->DrawObject(nid);
65 }
66 return 1;
67 }
In the CRIB processor, there is a processor that inherits from TModuleInfo, TModuleData.
In the constractor of this class use copy constractor of TModuleInfo, but the default artemis doesn’t implement it.
This class is used when we want to check the raw data.
For the detail, please see check raw data page.
Therefore, we modified this like this:
31 TModuleInfo::TModuleInfo(const TModuleInfo& rhs)
32+ : TParameterObject(rhs),
33+ fID(rhs.fID),
34+ fType(rhs.fType),
35+ fHists(nullptr)
36 {
37+ if (rhs.fHist) {
38+ fHists = new TObjArray(*(rhs.fHists));
39+ }
40+
41+ fRanges = rhs.fRanges;
42 }