Setting
This chapter describes how to prepare the configuration file for the experiment. If you have already some CRIB artemis environment, please see from here for initial settings.
This chapter describes how to prepare the configuration file for the experiment. If you have already some CRIB artemis environment, please see from here for initial settings.
If you installed with “curl” command explained previous chapter, you should have artnew command.
This command will make new experiment directory interactively.
Before using this command, please check and make the directory structure!
The word after “:” is your input.
> artnewcreate new artemis work directory? (y/n): y
Input experimental name: test
Is it OK? (y/n): y
Input value: testartnew: If no input is provided, the default value is used.
Input repository path or URL (default: https://github.com/okawak/artemis_crib.git):
Is it OK? (y/n): y
Input value: https://github.com/okawak/artemis_crib.gitInput rawdata directory path (default: /data/test/ridf):
Is it OK? (y/n): y
Input value: /data/test/ridfInput output data directory path (default: /data/test/user):
Is it OK? (y/n): y
Input value: /data/test/userBased on the repository, do you make your own repository? (y/n): y
is it local repository (y/n): y
artnew: making LOCAL repository of test
Input the local repository path (default: $HOME/repos/exp):
Is it OK? (y/n): y
Input value: /home/crib/repos/exp
-- snip --
art_analysis setting for test is finished!The initial setting is completed!!
After artnew command, you can see new directory of config files.
> tree -a art_analysis
art_analysis
├── .conf
│ ├── artlogin.sh
+│ └── test.sh
├── bin
│ ├── art_check
│ ├── art_setting
│ └── artnew
+└── test
This is experiment name “test” example.
In order to load this script test.sh, please modify “EXP_NAME” environment valiable in .zshrc.
export EXP_NAME="test" # your experimentAnd load the config file.
> source ~/.zshrcThen you can make your own work directory by using artlogin command!
For example, let’s make default user (user name is the same with experiment name)!
> artloginIf you want to make your own directory, the following will work.
> artlogin yournameartlogin: user 'test' not found.
create new user? (y/n): y
Cloning into '/Users/okawa/art_analysis/test/test'...
done.artlogin: making local git config
Input fullname: KodaiOkawa
Is it Okay? (y/n): y
Input email address: okawa@cns.s.u-tokyo.ac.jp
Is it Okay? (y/n): y> pwd
/home/crib/art_analysis/test/test
> ls -lIf your synbolic link seems okay, the setting is all!
If artnew setting have problem, the error message will appear.
Typical examples are as follows.
mkdir: /data: Read-only file systemThis is a case of the directory permissions not being set correctly.
Use the chmod command or similar to set them correctly and try again.
Before starting analysis, you need to build. The current version of the artemis use “cmake” so the following steps must be taken.
> artlogin (username)
> mkdir build && cd build
> cmake ..
> make -j4
> make install
> acdacd is the alias command that is definded after artlogin command. (acd = cd your_work_directory)
Also if you changed some processor, you need to do these process.
Then some important configuration files are automatically created.
> tree -L 1
.
+├── artemislogon.C
+├── thisartemis-crib.sh
-- snip --
└── run_artemis.cpp
Before starting artemis, you need to load the thisartemis-crib.sh.
The artlogin command is also used to read this script, so run this command again after the build.
> artlogin (usename)
> aThen you can start artemis by using a command!
Before configuring the settings according to your experiment, let’s check that artemis is working!
> artlogin (username)
> a # start the artemis!Then the prompt change to the artemis [0].
This means you are in artemis console!
Analysis using artemis use event loop.
It is therefore necessary to load a file that specifies what kind of analysis is to be performed.
This file is called the steering file.
As an example, let’s check the operation using a steering file that only generates random numbers!
The command to load the steering file is add.
artemis [0] add steering/example/example.tmpl.yaml NUM=0001 MAX=10This means that 10000 random numbers from 0 to MAX=10 are generated (10000 event loops). NUM=0001 is the ID, so any number is okay (related to outputed file name).
And the command to start the event loop is resume.
(Often abbreviated as “resume” or “re”.
The abbreviated form will also run without problems if there are no conflicts with other commands.)
artemis [1] res
artemis [2] Info in <art::TTimerProcessor::PostLoop>: real = 0.02, cpu = 0.02 sec, total 10000 events, rate 500000.00 evts/secWhen the time taken for such an analysis is displayed, it means that all event loops have been completed.
If you are doing a time-consuming analysis and want to suspend the event loop in the middle, suspend command is used. (Often “sus” or “su” is used.)
artemis [2] susThis event loop creates histogram objects (inherit from TH1 or TH2) and a TTree object. Let’s look at how to access each of these.
Details are given in the Histograms section, but histograms are created in an internal directory. To access it, you need to use the same commands as for the linux command, such as “ls” or “cd”, to get to that directory.
artemis [2] ls
artemis
> 0 art::TTreeProjGroup test2 test (2) # the first ">" means your current position
1 art::TTreeProjGroup test test
2 art::TAnalysisInfo analysisInfo
# then let's move to the "test" directory!
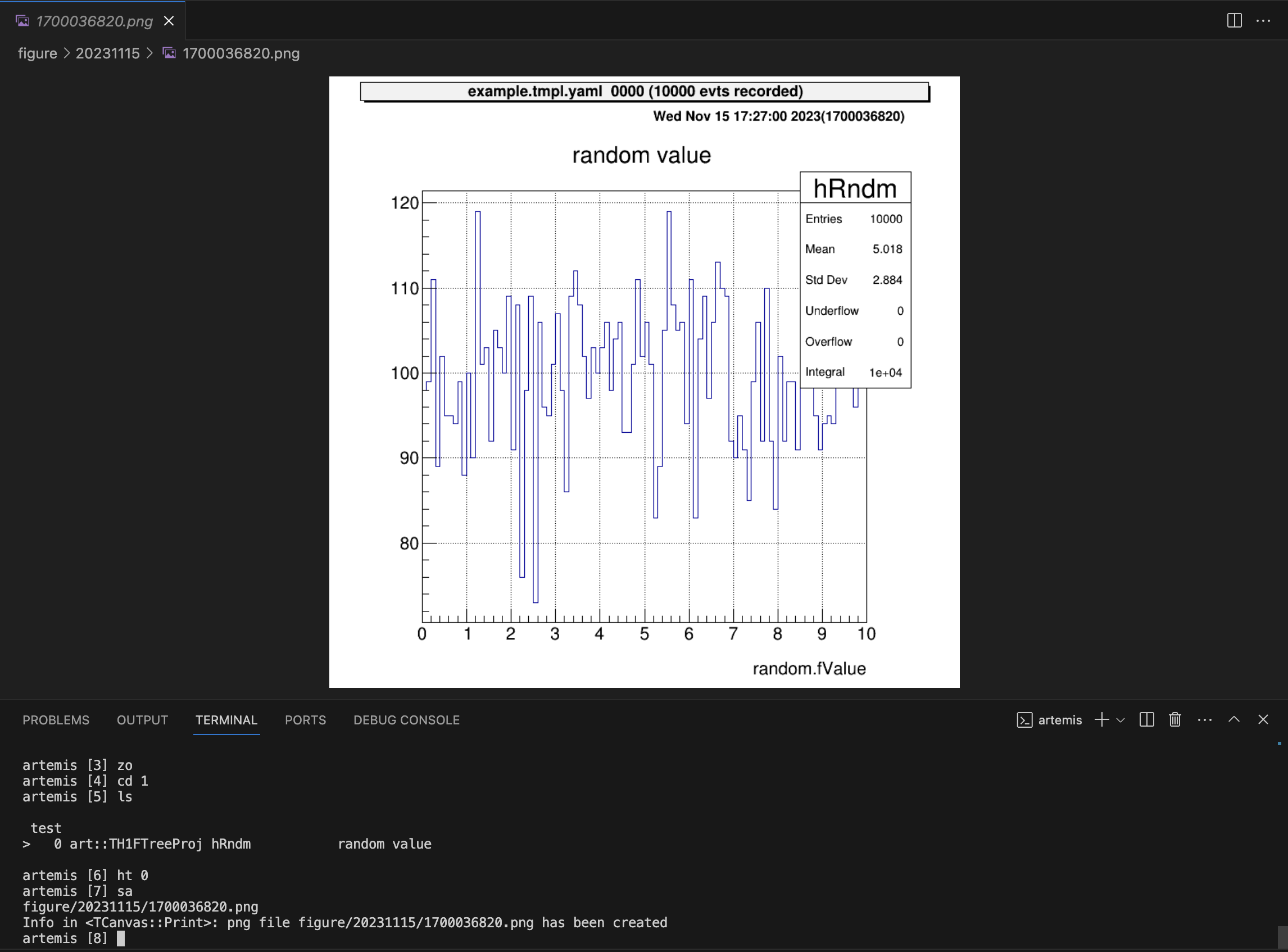
artemis [3] cd 1
artemis [4] ls
test
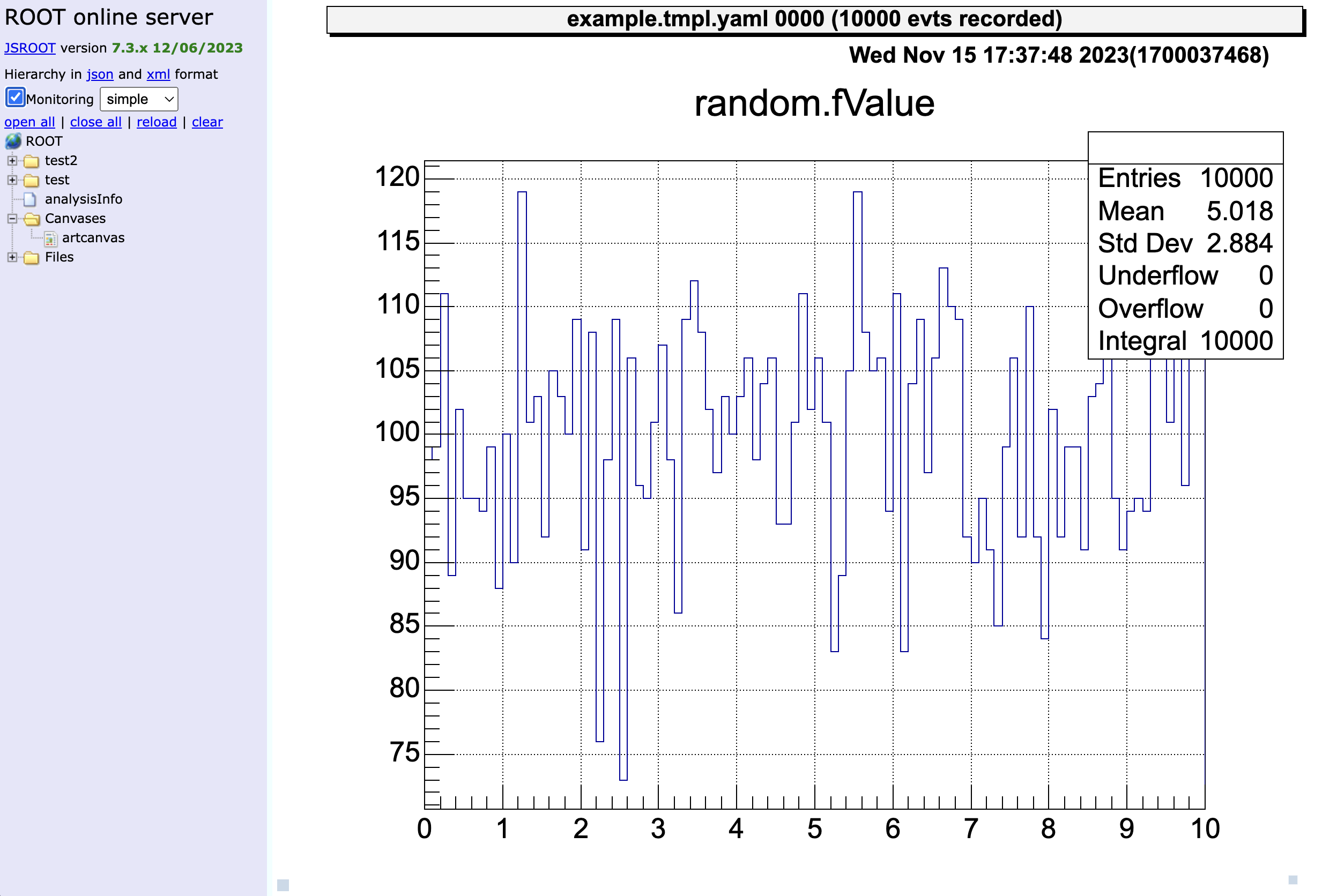
> 0 art::TH1FTreeProj hRndm random valueYou can use the command ht [ID] to display a histogram.
The ID can be omitted if it is already represented by >.
artemis [5] zone # make artemis canvas
artemis [6] ht 0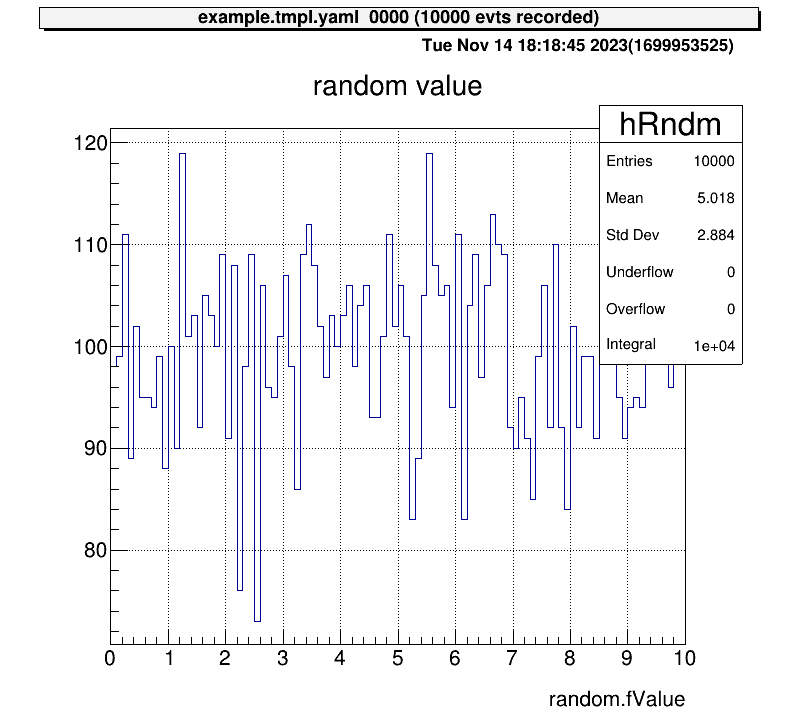
Next, let’s also check the histogram in “test2” directory and display two histograms vertically at the same time!
artemis [7] zone 2 1 # row=2, column=1
artemis [8] ht 0 # show the current hist
artemis [9] cd ..
artemis [10] ls
artemis
> 0 art::TTreeProjGroup test2 test (2)
1 art::TTreeProjGroup test test
2 art::TAnalysisInfo analysisInfo
artemis [11] cd 0
test2
> 0 art::TH1FTreeProj hRndm2 random number
artemis [12] ht 0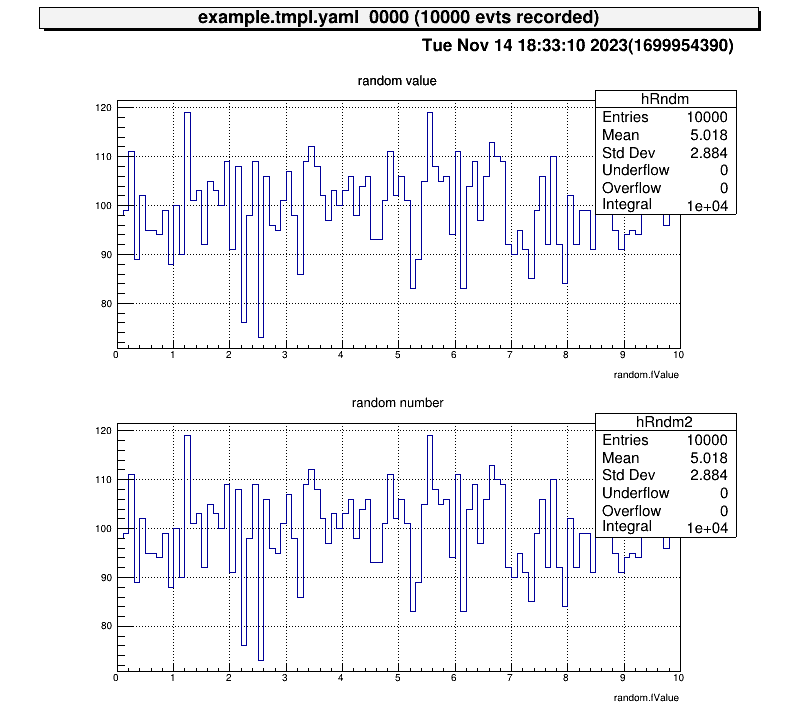
Now consider displaying a diagram from a TTree object. The file is created at here.
artemis [13] fls
files
0 TFile output/0001/example_0001.tree.root (CREATE)We use the fcd command to navigate to this root file.
artemis [14] fcd 0
artemis [15] ls
output/0001/example_0001.tree.root
> 0 art::TAnalysisInfo analysisInfo
1 art::TArtTree tree treeThe command branchinfo (“br”) displays a list of the branches stored in this tree.
artemis [16] br
random art::TSimpleDataAt the same time, the ROOT command can be used.
artemis [17] tree->Print()
******************************************************************************
*Tree :tree : tree *
*Entries : 10000 : Total = 600989 bytes File Size = 86144 *
* : : Tree compression factor = 7.00 *
******************************************************************************
*Br 0 :random : art::TSimpleData *
*Entries : 10000 : Total Size= 600582 bytes File Size = 85732 *
*Baskets : 1 : Basket Size= 3200000 bytes Compression= 7.00 *
*............................................................................*What is stored in the branch is not the usual type like “double” or “int”, but a class defined in artemis. Therefore, the “artemis” root file cannot be opened by usual ROOT.
Accessing data in a branch’s data class requires the use of public variables and methods, which can be examined by providing arguments to branchinfo [branch name] or classinfo [class name].
artemis [18] br random
art::TSimpleData
Data Members
Methods
Bool_t CheckTObjectHashConsistency
TSimpleData& operator=
TSimpleData& operator=
See also
art::TSimpleDataBase<double>
artemis [19] cl art::TSimpleDataBase<double>
art::TSimpleDataBase<double>
Data Members
double fValue
Methods
void SetValue
double GetValue
Bool_t CheckTObjectHashConsistency
TSimpleDataBase<double>& operator=
See also
art::TDataObject base class for data objectTherefore, it can be seen that it can be accessed by the value fValue.
artemis [20] zone
artemis [21] tree->Draw("random.fValue>>(100,0.,10.)")artemis [*] help # show the commands we can use
artemis [*] save # save the current canvas
artemis [*] print # print the current canvas (send to the printer, need to configure)
artemis [*] unzoom
artemis [*] lgy, lgz, lny, lnz # linear or log scaleFrom this section, we start to configure the settings according to the actual experimental setup. The setting files are followings:
> tree
.
├── mapper.conf
├── conf
│ ├── map
│ │ ├── ppac
│ │ │ ├── dlppac.map
-- snip --
│ └── seg
│ ├── modulelist.yaml
│ └── seglist.yaml
-- snip --The data obtained from an ADC/TDC is in the form of, for example, “The data coming into channel 10 of an ADC with an ID of 1 is 100”.
---
title: Data flow example
---
graph LR;
A{detector} -->|signal| B(TDC/ADC<br></br>ID=1, ch=10)
B -->|value=100| C[<strong>Data build</strong>\nridf file]
The role of the map file is to map this value of “100” to an ID that is easy to analyse. An ID that is easy to analyse means, for example, that even if signals from the same detector are acquired with different ADCs/TDCs, the same ID is easier to handle in the analysis.
---
title: Data flow example
---
graph LR;
A(TDC/ADC<br></br>ID=1, ch=10) -->|value=100| B[<strong>Data build</strong>\nridf file]
B -->|value=100 mapping to| C(analysis<br></br>ID=2, ch=20)
After mapping, we can check the data of this “100” from ID=2 and ch=20. This ID and channel (2, 20) are provided for convenience, so you can freely set them.
So, in summary, the map file role is like this:
---
title: role of the map file
---
graph LR;
A(DAQ ID<br></br>ID=1, ch=10) <-->|mapping| B(analysis ID<br></br>ID=2, ch=20)
CRIB is using Babirl for the DAQ system. In this system, the DAQ ID represented in the example is determined by five parameters.
The dev, fp, det and geo parameters can be set from DAQ setting. For the CRIB experiment, conventionally we set dev=12, fp=0–2 (for each MPV), det=6,7 (6=energy, 7=timing) and geo=from 0. But you can change it freely.
And analysis ID represented in the example is determined by two parameters.
Of cource you can also set the value freely.
The format of the map file is followings:
# [category] [id] [[device] [focus] [detector] [geo] [ch]] ....
1, 0, 12, 1, 6, 0, 0# map for SSD
# [category] [id] [[device] [focus] [detector] [geo] [ch]] ....
#
# Map: energy, timing
#
#--------------------------------------------------------------------
1, 0, 12, 1, 6, 0, 0, 12, 2, 7, 0, 0Please create map files for all detectors like this!
You can select the map file to be loaded with this file. This is especially useful when separating map files for testing from map files for the experiment.
The format is followings: (path/to/the/map/file number)
# file path for configuration, relative to the working directory
# (path to the map file) (Number of columns)
# cid = 1: rf
conf/map/rf/rf.map 1In the note example above, the number is 2.
Please do not forget to add to the mapper.conf after you add some map files.
This conf files are used when you use “chkseg.yaml” steering file. This steering file create raw data 2D histograms. I will describe in the Example: online_analysis/Check raw data in detail.
The steering file (yaml format) is a file that directs the process of how the obtained data is to be processed.
The artemis is an object-oriented program whose components are called processors, which are combined to process data.
The main role of the “processor” is to process data from an input data called InputCollection and create an output data called OutputCollection.
This “OutputCollection” will be stored into the root file as a “tree”.
Complex processing can be performed by using “processor” in multiple steps.
I will explain how to create this “steering” file using Si detector data as an example.
---
title: example of the data process structure
---
graph TD;
subgraph event loop
A-->B(mapping processor<br></br>InputCollection: decoded data\nOutputCollection: Si raw data)
B-->C(calibration processor<br></br>InputCollection: Si raw data\nOutputCollection: Si calibrated data)
C-->X((event end))
X-->A
end
subgraph DAQ
D(Raw binary data)-->A(decode processor<br></br>InputCollection: raw data\nOutputCollection: decoded data)
end
First, I describe what is the Anchor and how to use command line arguments.
See example here.
Anchor:
- &input ridf/@NAME@@NUM@.ridf
- &output output/@NAME@/@NUM@/chkssd@NAME@@NUM@.root
- &histout output/@NAME@/@NUM@/chkssd@NAME@@NUM@.hist.rootYou can use variables from elsewhere in the steering file by declaring them as such. For example if you write:
something: *inputThis unfolds as follows:
something: ridf/@NAME@@NUM@.ridfVariables enclosed in @ can also be specified by command line arguments.
For example, If you command like the following in the artemis console,
artemis [1] add steering/chkssd.yaml NAME=run NUM=0000it is considered as
Anchor:
- &input ridf/run0000.ridf
- &output output/run/0000/chkssdrun0000.root
- &histout output/run/0000/chkssdrun0000.hist.rootWhen using the “Babirl”, the data file will be in the form of “ridf”. In this case, the beginning and end of the steering file is usually as follows.
Processor:
- name: timer
type: art::TTimerProcessor
- name: ridf
type: art::TRIDFEventStore
parameter:
OutputTransparency: 1
InputFiles:
- *input
SHMID: 0
- name: mapper
type: art::TMappingProcessor
parameter:
OutputTransparency: 1
# -- snip --
- name: outputtree
type: art::TOutputTreeProcessor
parameter:
FileName:
- *outputEventStore (see below)OutputTransparency is set to 1, indicating that “OutputCollection” is not exported to the root file.
The “mapping processor” puts the data stored in the “EventStore” into a certain data class based on “mapper.conf”. Assume the following map file is used.
# map for SSD
# [category] [id] [[device] [focus] [detector] [geo] [ch]] ....
#
# Map: energy, timing
#
#--------------------------------------------------------------------
1, 0, 12, 1, 6, 0, 0, 12, 2, 7, 0, 0In this case, since we are assuming data from the Si detector, let’s consider putting it in a data class that stores energy and timing data, “TTimingChargeData”! The processor mapping to this data class is “TTimingChargeMappingProcessor”.
Processor:
- name: proc_ssd_raw
type: art::TTimingChargeMappingProcessor
parameter:
CatID: 1
ChargeType: 1
ChargeTypeID: 0
TimingTypeID: 1
Sparse: 1
OutputCollection: ssd_rawThen, you can access the ssd_raw data by using like tree->Draw("ssd_raw.fCharge")
While the data in the “ssd_raw” are raw channel of the ADC and TDC, it is important to see the data calibrated to energy and time. I will explain the details in Example: preparation/macro, but here I will discuss the calibration processors assuming that the following appropriate calibration files have been created.
Now, let’s load these files.
Processor:
- name: proc_ssd_ch2MeV
type: art::TParameterArrayLoader
parameter:
Name: prm_ssd_ch2MeV
Type: art::TAffineConverter
FileName: prm/ssd/ch2MeV.dat
OutputTransparency: 1
- name: proc_ssd_ch2ns
type: art::TParameterArrayLoader
parameter:
Name: prm_ssd_ch2ns
Type: art::TAffineConverter
FileName: prm/ssd/ch2ns.dat
OutputTransparency: 1To calibrate data contained in a TTimingChargeData class, a TTimingChargeCalibrationProcessor processor is used.
Processor:
- name: proc_ssd
type: art::TTimingChargeCalibrationProcessor
parameter:
InputCollection: ssd_raw
OutputCollection: ssd_cal
ChargeConverterArray: prm_ssd_ch2MeV
TimingConverterArray: prm_ssd_ch2nsNote here that “InputCollection”, “ChargeConverterArray”, and “TimingConverterArray” use the same names as the highlighted lines in the code block above.
The arguments to be used will vary depending on the processor used, so please check and write them according to the situation. If you want to check from artemis console, you can use “processordescription” command like this
> artlogin (username)
> a
artemis [0] processordescription art::TTimingChargeCalibrationProcessor
Processor:
- name: MyTTimingChargeCalibrationProcessor
type: art::TTimingChargeCalibrationProcessor
parameter:
ChargeConverterArray: no_conversion # [TString] normally output of TAffineConverterArrayGenerator
InputCollection: plastic_raw # [TString] array of objects inheriting from art::ITiming and/or art::ICharge
InputIsDigital: 1 # [Bool_t] whether input is digital or not
OutputCollection: plastic # [TString] output class will be the same as input
OutputTransparency: 0 # [Bool_t] Output is persistent if false (default)
TimingConverterArray: no_conversion # [TString] normally output of TAffineConverterArrayGenerator
Verbose: 1 # [Int_t] verbose level (default 1 : non quiet)If you want to analyse a large number of detectors, not just Si detectors, writing everything in one steering file will result in a large amount of content that is difficult to read.
In that case, we can use “include” node!
In the examples we have written so far, let’s only use a separate file for the part related to the analysis of the Si detector.
# -- snip --
Processor:
# -- snip --
- include: ssd/ssd_single.yaml
# -- snip --Processor:
# parameter files
- name: proc_ssd_ch2MeV
type: art::TParameterArrayLoader
parameter:
Name: prm_ssd_ch2MeV
Type: art::TAffineConverter
FileName: prm/ssd/ch2MeV.dat
OutputTransparency: 1
- name: proc_ssd_ch2ns
type: art::TParameterArrayLoader
parameter:
Name: prm_ssd_ch2ns
Type: art::TAffineConverter
FileName: prm/ssd/ch2ns.dat
OutputTransparency: 1
# data process
- name: proc_ssd_raw
type: art::TTimingChargeMappingProcessor
parameter:
CatID: 1
ChargeType: 1
ChargeTypeID: 0
TimingTypeID: 1
Sparse: 1
OutputCollection: ssd_raw
- name: proc_ssd
type: art::TTimingChargeCalibrationProcessor
parameter:
InputCollection: ssd_raw
OutputCollection: ssd_cal
ChargeConverterArray: prm_ssd_ch2MeV
TimingConverterArray: prm_ssd_ch2nsIn this way, the contents of “chkssd.yaml” can be kept concise, while the same process is carried out. Note that the file paths here are relative to the paths from the steering directory. Parameter files, for example, are relative paths from the working directory (one level down).
Utilising file splitting also makes it easier to check the steering files that analyse a large number of detectors like this.
# -- snip --
Processor:
# -- snip --
- include: rf/rf.yaml
- include: ppac/f1ppac.yaml
- include: ppac/dlppac.yaml
- include: mwdc/mwdc.yaml
- include: ssd/ssd_all.yaml
# -- snip --When you include other files, you can set arguments. This can be used, for example, to share variables. Details will be introduced in the example section.
The whole steering file is as follows:
Anchor:
- &input ridf/@NAME@@NUM@.ridf
- &output output/@NAME@/@NUM@/chkssd@NAME@@NUM@.root
- &histout output/@NAME@/@NUM@/chkssd@NAME@@NUM@.hist.root
Processor:
- name: timer
type: art::TTimerProcessor
- name: ridf
type: art::TRIDFEventStore
parameter:
OutputTransparency: 1
InputFiles:
- *input
SHMID: 0
- name: mapper
type: art::TMappingProcessor
parameter:
OutputTransparency: 1
- include: ssd/ssd_single.yaml
# output root file
- name: outputtree
type: art::TOutputTreeProcessor
parameter:
FileName:
- *outputProcessor:
# parameter files
- name: proc_ssd_ch2MeV
type: art::TParameterArrayLoader
parameter:
Name: prm_ssd_ch2MeV
Type: art::TAffineConverter
FileName: prm/ssd/ch2MeV.dat
OutputTransparency: 1
- name: proc_ssd_ch2ns
type: art::TParameterArrayLoader
parameter:
Name: prm_ssd_ch2ns
Type: art::TAffineConverter
FileName: prm/ssd/ch2ns.dat
OutputTransparency: 1
# data process
- name: proc_ssd_raw
type: art::TTimingChargeMappingProcessor
parameter:
CatID: 1
ChargeType: 1
ChargeTypeID: 0
TimingTypeID: 1
Sparse: 1
OutputCollection: ssd_raw
- name: proc_ssd
type: art::TTimingChargeCalibrationProcessor
parameter:
InputCollection: ssd_raw
OutputCollection: ssd_cal
ChargeConverterArray: prm_ssd_ch2MeV
TimingConverterArray: prm_ssd_ch2ns> acd
> a
artemis [0] add steering/chkssd.yaml NAME=run NUM=0000In the online analysis, it is important to have immediate access to data. The artemis can produce TTree object but long commands are needed to access, for example,
artemis [1] fcd 0 # move to the created rootfile
artemis [2] zone 2 2 # make a "artemis" 2x2 canvas
artemis [3] tree->Draw("ssd_cal.fCharge:ssd_cal.fTiming>(100,0.,100., 100,0.,100)","ssd_cal.fCharge > 1.0","colz")This would take time if there are some histograms that you want to display immediately…
Therefore, if you know in advance the diagram you want to see, it is useful to predefine its histogram!
The processor used is TTreeProjectionProcessor.
I would like to explain how to use this one.
Let’s look at how histograms are defined when looking at SSD data. First, let’s prepare the steering file as follows! please see previous section for omissions.
# -- snip --
- include: ssd/ssd_single.yaml
# Histogram
- name: projection_ssd
type: art::TTreeProjectionProcessor
parameter:
FileName: hist/ssd/ssd.yaml
Type: art::TTreeProjection
OutputFilename: *histout
# output root file
- name: outputtree
type: art::TOutputTreeProcessor
parameter:
FileName:
- *outputThe histogram is created based on the TTree object,
so describe the processing of the histogram after the part that represents the data processing and before the part that outputs the TTree (TOutputTreeProcessor).
There are three points to note here.
Therefore, I would now like to show the histogram definition file.
First please look at this example.
1anchor:
2 - &energy ["ssd_cal.fCharge",100,0.,100.]
3 - &timing ["ssd_cal.fTiming",100,0.,100.]
4alias:
5 energy_cut: ssd_cal.fCharge>1.0;
6group:
7 - name: ssd_test
8 title: ssd_test
9 contents:
10 - name: ssd_energy
11 title: ssd_energy
12 x: *energy
13
14 - name: ssd_timing
15 title: ssd_timing
16 x: *timing
17
18 - name: ssd_energy and timing
19 title: ssd_energy and timing
20 x: *timing
21 y: *energy
22 cut: energy_cutThis definition file consists of three parts.
The actual core part is the “2.3 group”, but “2.1 anchor” and “2.2 alias” are often used to make this part easier to write.
The anchor defines the first argument of tree->Draw("ssd_cal.fCharge>(100,0.,100.)","ssd_cal.fCharge > 1.0")
The array stored in the variable named “energy” in the second line looks like [str, int, float, float] and has the following meanings
As you might imagine, inside the first argument you can also add operations such as TMath::Sqrt(ssd_cal.fCharge) or ssd_cal.fCharge-ssd_cal.fTiming, because it is the same with “tree->Draw”.
Note, however, that the definition here is for one-dimensional histograms. Two-dimensional histograms will be presented in Section 2.3. It is very simple to write!
This part is used when applying gates to events (often we call it as “cut” or “selection”).
For example, if you only want to see events with energies above 1.0 MeV, you would write something like tree->Draw("energy","energy>1.0").
The alias node is used to define the part of energy>1.0
A semicolon “;” at the end of the sentence may be needed…? please check the source.
The histogram is defined here and the object is stored in a directory in artemis (ROOT, TDirectory). In the example shown above, the directory structure would look like this:
(It is not actually displayed in this way).
# in artemis
.
└── ssd_test
├── ssd_energy (1D hist)
├── ssd_timing (1D hist)
└── ssd_energy and timing (2D hist)The first “name” and “title” nodes are arguments of TDirectory instance. Also the second “name” and “title” nodes are arguments of instance of TH1 or TH2 object. The other “x”, “y” and “cut” is the important node!
There are many useful command for checking the histogram objects. These are similar to the ANAPAW commands.
> artlogin (username)
> a
artemis [0] add steering/chkssd.yaml NAME=hoge NUM=0000
artemis [1] res
artemis [2] sus
artemis [3] ls # check the artemis directory
artemis
> 0 art::TTreeProjGroup ssd_test ssd_test
1 art::TAnalysisInfo analysisInfoartemis [4] cd 0
artemis [5] ls
ssd_test
> 0 art::TH1FTreeProj ssd_energy ssd_energy
1 art::TH1FTreeProj ssd_timing ssd_timing
2 art::TH2FTreeProj ssd_energy and timing ssd_energy and timingartemis [6] ht 0
artemis [7] ht 2 colzWhen setting up several detectors of the same type and wanting to set up a histogram with the same content, it is tedious to create several files with only the names of the objects changed. In such cases, it is useful to allow the histogram definition file to have arguments.
Please look here first.
# -- snip --
- include: ssd/ssd_single.yaml
# Histogram
- name: projection_ssd
type: art::TTreeProjectionProcessor
parameter:
FileName: hist/ssd/ssd.yaml
Type: art::TTreeProjection
OutputFilename: *histout
Replace: |
name: ssd_cal
# -- snip --We add the highlighted lines.
Then the “name” can be used at hist file by @name@!
The “name” can be freely set.
anchor:
- &energy ["@name@.fCharge",100,0.,100.]
- &timing ["@name@.fTiming",100,0.,100.]
alias:
energy_cut: @name@.fCharge>1.0;
group:
- name: ssd_test
title: ssd_test
contents:
- name: ssd_energy
title: ssd_energy
x: *energy
- name: ssd_timing
title: ssd_timing
x: *timing
- name: ssd_energy and timing
title: ssd_energy and timing
x: *timing
y: *energy
cut: energy_cutThis is useful when there are more objects to check!
# -- snip --
- include: ssd/ssd_single.yaml
# Histogram
- name: projection_ssd
type: art::TTreeProjectionProcessor
parameter:
FileName: hist/ssd/ssd.yaml
Type: art::TTreeProjection
OutputFilename: *histout
Replace: |
name: ssd_cal
- name: projection_ssd
type: art::TTreeProjectionProcessor
parameter:
FileName: hist/ssd/ssd.yaml
Type: art::TTreeProjection
Replace: |
name: ssd_raw
# and so on!
# -- snip --File splitting using “include” nodes, as described in the section on steeling, can also be used in the same way.
When we start the analysis, there are many situations where the analysis server on which artemis is installed is not only operated directly, but also remotely using “ssh”.
In such cases, there are various settings that need to be made in order for the figure to be displayed on the local computer, and some of these methods are described in this section.
We recommended to use VNC server currently, but note that policies may change in the future.
This is a list of ways to display the figures.
This is the simplest method. Simply transfer the remote X to the local.
ssh -X analysisPCThis “X” option allow the X11Forwarding.
However, the problem with this method is that it takes a long time to process, and it takes longer from the time the command is typed until it is drawn. It is also not recommended as the process can become slow if a large number of people use it at the same time.
However, it is simpler than other methods and should be used when necessary, e.g. for debugging.
This is old version of VNC server (TigerVNC). Latest version supports more secure method, so this method may no longer be avaliable in the future…
First please install VNC viewer to your PC. Any viewer may work well, but we are using this software.
First, please check the ID number of the VNC server we are running.
> vncserver -list
TigerVNC server sessions:
X DISPLAY # PROCESS ID
:1 3146
:5 7561
:2022 29499
:2 23055In this example, number 1, 5, 2022 and 2 VNC server is running. And select an available number to start the VNC server you want to use.
> vncserver :10 # start the VNC server!If you want to kill the VNC server, the below command will work.
> vncserver -kill :10 # kill the VNC server!Next, configure the canvas created by artemis to be sent to a VNC server.
The a command can treat this process by using .vncdisplay file!
> artlogin (username) # move to your artemis work directory
> echo "10" > .vncdisplay # write the ID of VNC server to the .vncdisplay fileThen, the setting in analysis PC is completed! The next step is to set up your local PC to receive this.
If you connect your PC in the same network with analysis PC, you can directory connect using the VNC viewer. However, CRIB analysis PC are connected CNS local network. In order to connect from outside network, we need to use “CNS login server”. If you want to make the login server account, please contact the CRIB member!
In this section, we are assuming that you have a CNS login server account.
To access the analysis PC, use two-stage ssh. Prepare the following configuration file.
Host login
# need to change
HostName CNS_loginserver_hostname
User username
IdentityFile ~/.ssh/id_rsa
# no need to change (if you want)
ForWardX11Timeout 24h
ControlPersist 30m
ForwardAgent yes
ControlMaster auto
ControlPath ~/.ssh/mux-%r@%h:%p
# any name is okay
Host analysis
# need to change
HostName analysisPC_hostname
User username
IdentityFile ~/.ssh/id_rsa
# no need to change (if you want)
ProxyCommand ssh login nc %h %p
ForwardAgent yes
ControlMaster auto
ControlPath ~/.ssh/mux-%r@%h:%p
ControlPersist 30mThen you can access to the analysis PC simply by:
> ssh analysisNext, in order to receive from the VNC server, we use port-forwarding! VNC servers with ID x use port number 5900+x. For example if we use number “10”, the port will be 5910.
Forward this to a certain port on localhost. This number can be any number that is not in use.
---
title: An example of port-forwarding
---
graph LR;
A(analysis PC<br></br>port 5910) --> |send|B(local PC<br></br>port 55910)
Host analysis
HostName analysisPC_hostname
User username
IdentityFile ~/.ssh/id_rsa
LocalForward 55910 localhost:5910
ProxyCommand ssh login nc %h %p
ForwardAgent yes
ControlMaster auto
ControlPath ~/.ssh/mux-%r@%h:%p
ControlPersist 30mThis allows you to display a VNC display by accessing port 55910 on your own PC (localhost), instead of having to access port 5910 on the analysis PC!
If your PC is in the same network, changing “localhost” to the “IP address of analysis PC” is okay (ex. 192.168.1.10:5910).
VScode is very exciting editor! The extension supports ssh and allows remote png files to be displayed on the editor.
However, it is a bit time-consuming as the diagram has to be saved each time to view it. Please refer to this as one method.
This is option…
Now the histogram object cannot display by JSROOT, because the object is not actually “TH1” or “TH2” object but “TH1FTreeProj” or “TH2FTreeProj”. (ref: issue#40)
We can only display the “TCanvas” Object.