Example
Up to now, we have introduced the installation and concepts of artemis. This chapter will show you how to analyse with artemis through practical examples; if you want to know how to use artemis, it is no problem to start reading from here.
Up to now, we have introduced the installation and concepts of artemis. This chapter will show you how to analyse with artemis through practical examples; if you want to know how to use artemis, it is no problem to start reading from here.
This section explain the example of preparation by using some useful macro.
I would now like to introduce the actual analysis using the CRIB analysis server. There are two ways to enter the analysis server, directly or remotely via ssh. If you come to CRIB and operate the server directly, I think it is quicker to analyse using the server while asking the CRIB members directly as I think they are nearby.
To enter the CRIB server, you need to enter the CNS network. To do this, you need to create an account on the CNS login server. Please contact Okawa (okawa@cns.s.u-tokyo.ac.jp) or the person responsible for CRIB (see here) and tell us that you want a login server account.
The CNS login server uses public key cryptography, so you need to send a shared key when you apply. This section describes how to create the key, especially on MacOS.
cd # move to /Users/yourname/ (home directory)
mkdir .ssh # if there is no .ssh directory
cd .ssh
ssh-keygenYou will be asked a number of interactive questions after this command, all of which are fine by default (Enter). Then you will see the pair of public-key and private-key.
ls
id_rsa id_rsa.pubid_rsa is the private-key, and id_rsa.pub is the public-key.
The private key is important for security reasons and should be kept on your own computer.
Then, please send this public-key to the CNS member.
in MacOS, open . command will open a finder for that directory, so it is easy to attach it to an email from here.
In the email,
are needed.
Next, let’s set up multi-stage ssh. As the login server is just a jump server, it is useful to be able to ssh to the CRIB analysis server at once! So create the following config file. The file placed in this directory is automatically read when you ssh.
cd ~/.ssh
vi config 1Host login
2 HostName CNS_loginserver_hostname
3 User username
4 IdentityFile ~/.ssh/id_rsa
5 ForWardX11Timeout 24h
6 ControlPersist 30m
7 ForwardAgent yes
8 ControlMaster auto
9 ControlPath ~/.ssh/mux-%r@%h:%p
10
11# any name is okay
12Host cribana
13 HostName analysisPC_hostname
14 User crib
15 IdentityFile ~/.ssh/id_rsa
16 ProxyCommand ssh login nc %h %p
17 ForwardAgent yes
18 ControlMaster auto
19 ControlPath ~/.ssh/mux-%r@%h:%p
20 ControlPersist 30mYou will be informed of the second and third lines above that we highlighted, so please change this parts. And ask the IP address of the CRIB analysis PC to the CRIB member, and change the 13 line.
Then you can enter the CRIB analysis PC just by
ssh cribanaCRIB member will tell you the passward!
For the VNC server (local forwarding), please see this section.
When you enter the CRIB computer, please check this is zsh shell.
> echo $SHELL
/usr/local/bin/zshCurrently, zsh installed locally is used. It is planned to update the OS in the future, after which it will differ from this path in the future.
If it is not zsh (like bash), please command
> zshThen you can start to configure by
> artlogin yourname
# input your information...
> mkdir build
> cd build
> cmake ..
> make -j4
> make install
> acdFor the detail, please check here.
> acd # move to your artemis work directory
> a # start artemis!
> a macro/macro.C # run macro script# read steering file
artemis [*] add steering/hoge.yaml NAME=hoge NUM=0000
# start event loop
artemis [*] res
artemis [*] start # defined in CRIB artemis
# stop event loop
artemis [*] sus
artemis [*] stop # defined in CRIB artemis
# help
artemis [*] help
# quit from artemis
artemis [*] .q# check and move the directory
artemis [*] ls
artemis [*] cd 0 # cd ID
# move to home directory in artemis
artemis [*] cd # cd .. will work?
# draw the histograms
artemis [*] ht 0 colz # ht ID option
artemis [*] hn colz # draw the next histogram object
artemis [*] hb colz # draw the previous histogram object
# divide the canvas
artemis [*] zone 2 2 # 2 x 2 canvas
# save and print the canvas
artemis [*] sa
artemis [*] pri# check the files
artemis [*] fls
# move to the created ROOT file
artemis [*] fcd 0 # fcd fileID
# check the all branches
artemis [*] br
# check the data members or methods
artemis [*] br branchname # ex. artemis [1] br ppaca
# the name of TTree object is "tree" (actually TArtTree object)
artemis [*] tree->Draw("ppaca.fY:ppaca.fX>>ppaca(100,-20.,20., 100,-20.,20.)","","colz")See here for an example using random numbers.
CRIB uses a multi-hit TDC called V1190 to take timing data (manual). When a trigger comes into this module, it opens a window with a set time width and records the timing of the data.
However, even if the signal is sent at exactly the same time to the trigger, due to the uncertainty in opening that window, the resulting channel will vary. Since absolute channel values will vary, but relative channel values for a given (especially trigger) timing will remain the same, it is necessary to subtract all data by some reference channel to achieve good timing resolution.
The signal that serves as the reference for that time is what we call Tref!
(Time reference)
Since it is essential that all events contain that data, we put the trigger signal in one of the channels and make it a Tref.
The “tref” settings are made in the following file:
Processor:
# J1 V1190A
- name: proc_tref_v1190A_j1
type: art::TTimeReferenceProcessor
parameter:
# [[device] [focus] [detector] [geo] [ch]]
RefConfig: [12, 2, 7, 0, 15]
SegConfig: [12, 2, 7, 0]Parameters RefConfig and SegConfig are set using the same ID as in the map file.
The “RefConfig” represents the “Tref” signal and the “SegConfig” represents the V1190 module. Therefore, the role of the processor is to subtract the “Segconfig” V1190 all timing signal from the “RefConfig” tref signal.
To apply this processor, add the following line to the steering file. For example,
Anchor:
- &input ridf/@NAME@@NUM@.ridf
- &output output/chkf3@NAME@@NUM@.root
- &histout output/chkf3@NAME@@NUM@.hist.root
Processor:
- name: timer
type: art::TTimerProcessor
- name: ridf
type: art::TRIDFEventStore
parameter:
OutputTransparency: 1
InputFiles:
- *input
- name: mapper
type: art::TMappingProcessor
parameter:
OutputTransparency: 1
- include: tref.yaml
- include: rf/rf.yaml
- include: coin/coin.yaml
- include: ppac/dlppac.yaml
- include: ssd/f3ssd.yaml
- name: outputtree
type: art::TOutputTreeProcessor
parameter:
FileName:
- *outputThe tref.yaml should be written before the main processor. In this example, it is written right after TMappingProcessor, and we recommend writing it in this position.
CRIB use two kinds of PPAC (Parallel-Plate Avalanche Counter), charge division method or delay-readout method. The PPAC placed at the F1 focal plane is charge-devision type, and the parameters to be converted to position are fixed and do not need to be calibrated. Therefore we explain the calibration method for delay-line PPAC (dl-PPAC).
Here we briefly describe the principle of converting from the obtained signal to position, but for more details, see here1.
We will discuss the x-direction because x and y is exactly same. First, define the parameters as follows
The artemis codes calculate the X position like this formula (see TPPACProcessor.cc).
$$ X~\mathrm{[mm]} = k_x\left( \frac{T_{X1} - T_{X2} + T_{Xin-offset} - T_{Xout-offset}}{2} \right) - X_{offset}$$Check the sign carefully! We often mistook the direction!!
The $T_{X1},~T_{X2}$ are measured value by TDC, and $k_x$ and $T_{Xin-offset}$ are specific value to PPAC, so we need to care only $T_{Xout-offset}$ and $X_{offset}$. $X_{offset}$ value depends on where we put the PPAC, so what we have to do is determine the line calibration parameter ( $T_{Xout-offset}$).
The following is a list of dl-PPAC parameters used in CRIB experiment.
| PPAC ID | $k_x$ [mm/ns] | $k_y$ [mm/ns] | $T_{Xin-offset}$ | $T_{Yin-offset}$ |
|---|---|---|---|---|
| #2 | 1.256 | 1.256 | 0.29 mm | 0.18 mm |
| #3 | 1.264 | 1.253 | 0.22 mm | 0.30 mm |
| #7 | 1.240 | 1.242 | 0.92 ns | 1.58 ns |
| #8 | 1.241 | 1.233 | 0.17 ns | 0.11 ns |
| #9 | 1.257 | 1.257 | 0.05 mm | 0.04 mm |
| #10 | 1.257 | 1.257 | 0.05 mm | 0.04 mm |
Different units are used for the offset. However, since the effect of this offset is eventually absorbed to the other offset value, it is no problem to use the values if we calibrate it correctly.
PPAC parameters are defined in the following files
For example, it is like this:
Type: art::TPPACParameter
Contents:
# #7 PPAC
f3bppac: # this is the name of PPAC, should be the same name with the one in steering file!
ns2mm:
- 1.240
- 1.242
delayoffset:
- 0.92
- 1.58
linecalib:
- 1.31
- -1.00
# 0: no exchange, 1: X -> Y, Y -> X
exchange: 0
# 0: no reflect, 1: X -> -X
reflectx: 1
geometry:
- 0.0
- 0.5
- 322.0
TXSumLimit:
- -800.0
- 2000.0
TYSumLimit:
- -800.0
- 2000.0ns2mm
This is $k_x$ and $k_y$ parameters -> input the fixed value
delayoffset
This is $T_{Xin-offset}$ and $T_{Yin-offset}$ parameters -> input the fixed value
linecalib
This is explained next.
exchange, reflectx
This parameter should be changed depending on the direction in which the PPAC is placed. The meanings of the parameters are given above as comments.
CRIB takes a coordinate system such that when viewed from downstream of the beam, the x-axis is rightward and the y-axis is upward. In other words, it takes a right-handed coordinate system with the beam as the Z-axis. While looking at the actual data, change these parameters so that the coordinate system becomes this coordinate system.
geometry
In the Line calibration, please set this value to (0,0). After Line calibration, if we put the PPAC with some geometry offset, we should change this parameters. Please be careful that the parameter will add minus this value for X and Y. Z offset will be used for TPPACTrackingProcessor.
TXSumLimit, TYSumLimit
Used to determine if it is a good event or not. Currently not used.
Before starting line calibration, please make sure that map file and steering file is correctly set. Also we need parameter file of prm/ppac/ch2ns.dat to convert TDC channel to ns unit. (already prepared I think)
graph LR;
A[TDC channel] -->|prm/ppac/ch2ns.dat| B[ns scale]
B --> |prm/ppac/dlppac.yaml|C{PPAC object}
When you complete the setting except for linecalib parameters, let’s start calibration!
We prepared two useful macros to calibrate dl-PPAC.
First, we have to prepare the data with masks on the PPAC like following picture. This mask has holes at 12.5 mm intervals.

The position of the alpha line through the central hole can be calculated and the offset adjusted to achieve that position. The geometry inside the PPAC is as follows. I think all PPAC geometries used by CRIB are the same.
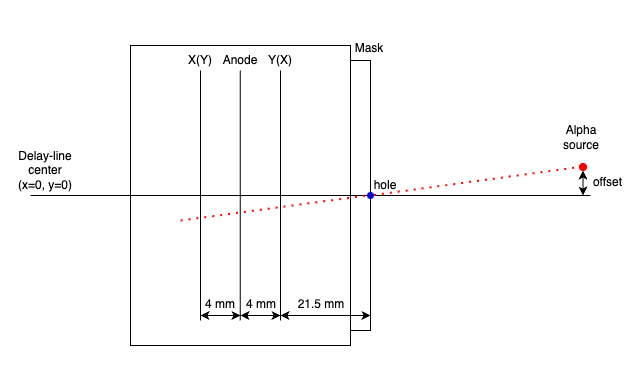
The parameters required to calculate the coordinates of the position are as follows.
Using these parameters, the macro/PPACLineCalibration.C calculate the theoretical position and how much the parameters should be moved.
Let’s calibrate PPACa as an example.
When we set the parameter like this files, the XY figure can be obtained.
Type: art::TPPACParameter
Contents:
# #2 PPAC
f3appac:
ns2mm:
- 1.256
- 1.256
delayoffset:
- 0.29
- 0.18
linecalib:
- 0.0
- 0.0
exchange: 0
reflectx: 1
geometry:
- 0.0
- 0.0
- -677.0 # user defined
TXSumLimit:
- -800.0
- 2000.0
TYSumLimit:
- -800.0
- 2000.0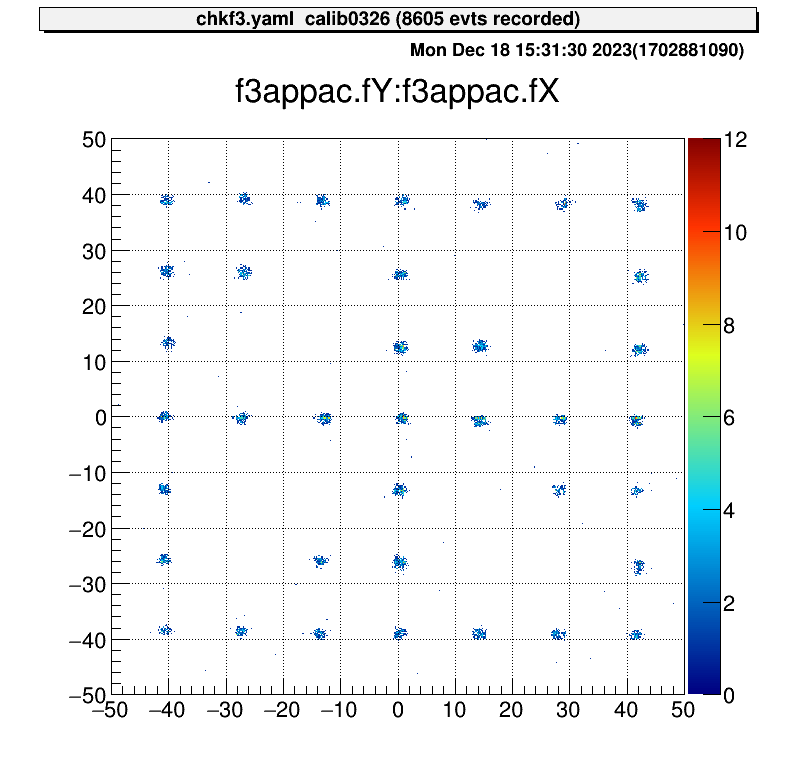
Then we can run the macro.
> acd
> vi macro/run_PPACLineCalibraion.C
# please set the parameters.
# instruction is written in this file
> a
artemis [0] add steering/hoge.yaml NAME=hoge NUM=0000
artemis [1] res
artemis [2] .x macro/run_PPACLineCalibration.C
# -- snip --
===================================================
center position (cal) : (-0, -0.51413)
center position (data) : (0.890109, -0.274066)
difference : (0.890109, 0.240065)
move parameters : (-1.41737, 0.382269)
===================================================And please input this value to the dlppac.yaml.
Type: art::TPPACParameter
Contents:
# #2 PPAC
f3appac:
ns2mm:
- 1.256
- 1.256
delayoffset:
- 0.29
- 0.18
linecalib:
- -1.417
- 0.382
exchange: 0
reflectx: 1
geometry:
- 0.0
- 0.0
- -677.0 # user defined
TXSumLimit:
- -800.0
- 2000.0
TYSumLimit:
- -800.0
- 2000.0Then you can complete the line calibration of the PPAC.
> a
artemis [0] add steering/hoge.yaml NAME=hoge NUM=0000
artemis [1] res
artemis [2] .x macro/run_PPACLineCalibration.C
# -- snip --
===================================================
center position (cal) : (-0, -0.51413)
center position (data) : (-0.0191028, -0.571067)
difference : (-0.0191028, -0.0569366) # <= almost zero!
move parameters : (0.0304184, -0.0906633)
===================================================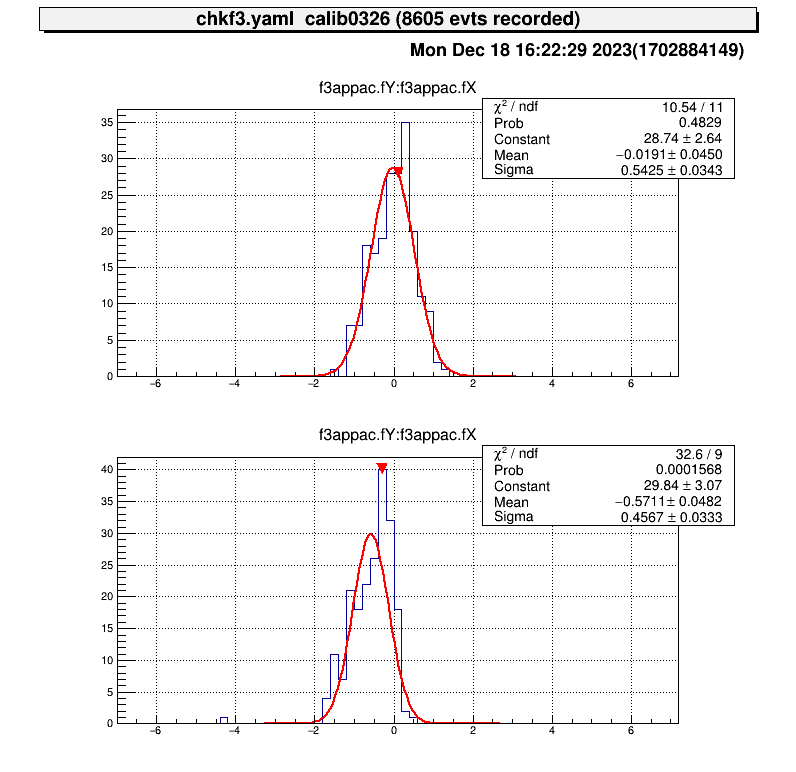
Because of the accuracy of the fitting, it does not make much sense to move the parameters any further.
The PPAC is then ready to be used by measuring how much offset the beamline axis has with respect to the delay-line axis at the position where the PPAC is actually placed, and putting this into the geometry parameters.
H. Kumagai et al., Nucl. Inst. and Meth. A 470, 562 (2001) ↩︎
updating…
This is the CRIB alpha source information. (unit: MeV)
| ID | alpha-2 | alpha-3 |
|---|---|---|
| 4.780 | 3.148 | |
| 5.480 | 5.462 | |
| 5.795 | 5.771 |
SSD calibration files need to be set at prm/ssd/ directory.
The directory structure is like this:
$ tree -L 2 prm/ssd
prm/ssd
├── ch2MeV.dat # test file
├── ch2ns.dat # test file
├── f2ch2MeV.dat
├── f2ch2MeV_raw.dat
├── f2ch2ns.dat
├── tel1
│ ├── ch2MeV_dEX.dat
│ ├── ch2MeV_dEX_raw.dat
│ ├── ch2MeV_dEY.dat
│ ├── ch2MeV_dEY_raw.dat
│ ├── ch2MeV_E.dat
│ ├── ch2MeV_E_raw.dat
│ ├── ch2ns_dEX.dat
│ ├── ch2ns_dEY.dat
│ ├── ch2ns_E.dat
│ └── tel_conf.yaml # telescope configuration, explain later
-- snip --The prm/ssd/ch2MeV.dat and prm/ssd/ch2ns.dat are used for test, so in the beam time measurement, actually this files are not necessory.
And prm/ssd/f2* files are used for F2SSD calibration, and files in prm/ssd/tel1/ directory are used for SSDs of a telescope.
The ch2ns.dat depends on TDC setting, so basically we don’t have to care so muc
(Usually the setting (the value) is same with previous experiment.), so we have to prepare the ch2MeV.dat files!
The file name need to be set like this example. The loaded parameter file name is defined SSD steering file, and we don’t want to change the SSD steering files so much, so please use such file names.
The “ch2MeV.dat” file format is like this:
# offset gain
1.7009 0.0173
0.0 1.0 # if there are some SSDs or strip SSD, you can add the line.We prepared useful macros to calibrate many SSDs. Please check these for more information.
It is sufficient to use the AlphaCalibration.C, but it is recommended to use the run_AlphaCalibration.C to keep a record of what arguments were used to calibrate.
After you prepared alpha calibration data and steering file (for example steering/calibration.yaml) to show raw data, you can use this macro.
$ acd
$ vi macro/run_AlphaCalibration.C
# please set the parameters.
# instraction is written in this file
$ a
artemis [0] add steering/hoge.yaml NAME=hoge NUM=0000
artemis [1] res
artemis [2] .x macro/run_AlphaCalibration.CThen the parameter file that defined at the “run_AlphaCalibration.C” and calibration figures will be created automatically.
These are example of the figures;

$ a
artemis [0] add steering/hoge.yaml NAME=hoge NUM=0000
artemis [1] res
artemis [2] fcd 0
artemis [3] zo
artemis [4] tree->Draw("...") # draw calibrated data
artemis [5] gStyle->SetOptStat(0)
artemis [6] sa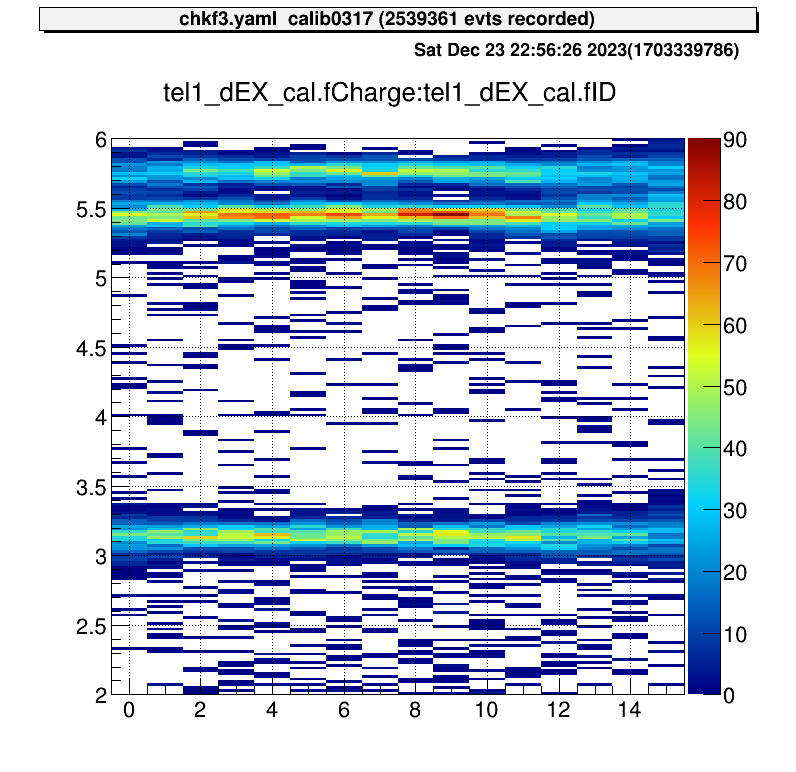
updating…
updating…
Analysis files for each experiment are managed using git.
This is so that they can be quickly restored if they are all lost for some reason.
Git is a bit complicated and you can commit freely if you are knowledgeable, but if you are unfamiliar with it, you don’t have to worry too much.
The main use is that if someone creates a useful file, it will be reflected for each user as well.
Here is a brief description of how to use it.
In the CRIB analysis PC, we used local repository. The files related the repository is stored here.
> cd ~
> tree -L 1 repos/exp
repos/exp
├── he6p2024.git
├── he6p.git
└── o14a.git
# 2023/12/18 current statusNote that if you delete the files in this directory, you will lose all backups.
I will describe the most commonly used commands and how to resolve conflicts.
This section explain the example of the online analysis in the CRIB experiment.
From here, we would like to explain in detail how to analyze the actual experiment. We assume that you have already prepared your the analysis environment. It is okay either your own directory or the default directory (see CRIB configuration). If you are not ready yet, please see here.
So let’s start to check the data.
At the F1 focal plane, there is (charge-divition) PPAC.
The steering file to analyze f1 data is chkf1.yaml.
We usually use “chk” (check) as a prefix of the steering files to analyze from raw binaly data.
$ artlogin (username)
$ a
artemis [0] add steering/chkf1.yaml NAME=hoge NUM=0000
artemis [1] resThe important data is “X position” at the F1PPAC. The histogram can be check by following step:
artemis [2] sus
artemis [3] ls
artemis
> 0 art::TTreeProjGroup f1check f1_check
1 art::TAnalysisInfo analysisInfo
artemis [4] cd 0
artemis [5] ls
f1check
> 0 art::TH1FTreeProj f1ppac_X f1ppac X
1 art::TH1FTreeProj f1ppac_Y f1ppac Y
2 art::TH1FTreeProj f1ppac_X1raw f1ppac X1raw
3 art::TH1FTreeProj f1ppac_X2raw f1ppac X2raw
4 art::TH1FTreeProj f1ppac_Y1raw f1ppac Y1raw
5 art::TH1FTreeProj f1ppac_Y2raw f1ppac Y2raw
6 art::TH2FTreeProj f1ppac_x_y f1ppac X vs Y
7 art::TH2FTreeProj f1ppac_x_rf f1ppac X vs RF
8 art::TH2FTreeProj f1ppac_x1_x2 f1ppac x1 vs x2
9 art::TH2FTreeProj f1ppac_y1_y2 f1ppac y1 vs y2Many histograms are defined, but in practice it’s enough to check the first X position. Sometimes we check other raw data histograms to see if the behavior of F1PPAC is correct or not.
artemis [6] ht 0Usually a gaussian fit is performed to get center the position.
artemis [7] xfFor the commane “xf” (xfitg), please check here.
When you think the signals from F1PPAC is okay, but position seems wrong (the X position is different from the setting of F1 slit), pleace modify the parameter files.
It is actually charge-divition PPAC, but the structure of parameter file is the same with dl-PPAC, so please also check PPAC preparation.
The basic usage is the same.
We use this steering file to check F2 data.
$ artlogin (username)
$ a
artemis [0] add steering/chkf2.yaml NAME=hoge NUM=0000
artemis [1] res
artemis [2] sus
artemis [3] ls
artemis
> 0 art::TTreeProjGroup f2check f2_check
1 art::TAnalysisInfo analysisInfoartemis [4] cd 0
artemis [5] ls
f2check
> 0 art::TH1FTreeProj f2PPAC X f2ppac x
1 art::TH1FTreeProj f2PPAC Y f2ppac y
2 art::TH1FTreeProj f2SSD raw f2ssd raw
3 art::TH1FTreeProj f2SSD raw (low) f2ssd raw (low)
4 art::TH1FTreeProj f2SSD cal f2ssd cal
5 art::TH2FTreeProj f2PPAC X vs Y f2ppac X vs Y
6 art::TH2FTreeProj F2PPAC X vs RF0 f2ppac X vs rf0
7 art::TH2FTreeProj F2PPAC Y vs RF0 f2ppac Y vs rf0
8 art::TH2FTreeProj RF0 vs F2SSD raw rf0 vs f2ssd raw
9 art::TH2FTreeProj RF0 vs F2SSD cal rf0 vs f2ssd cal
10 art::TH2FTreeProj RF1 vs F2SSD cal rf1 vs f2ssd cal
11 art::TH2FTreeProj F2PPAC X vs F2SSD raw f2ppac x vs f2ssd raw
12 art::TH2FTreeProj F2PPAC X vs F2SSD cal f2ppac x vs f2ssd cal
13 art::TH2FTreeProj F2PPAC Y vs F2SSD cal f2ppac y vs f2ssd cal
14 art::TH1FTreeProj RF0 rf0As you know, you can check the histograms
# for 1D histograms
artemis [*] ht [id]
artemis [*] hn # histogram next
artemis [*] hb # histogram before (back?)
# for 2D histograms
artemis [*] ht [id] colz # colz is option
artemis [*] hn colz
artemis [*] hb colzIf you want to save,
artemis [*] sa
artemis [*] priupdating…
updating…
In the CRIB experiment, we often use a “telescope” consisting of DSSSD (Double-Sided SSD) and SSD (Single-Pad SSD).
The combination of these multiple Si detectors as a dE-E detector is called a telescope.
To analyze the data as telescope data rather than individual detectors, I created a data class called TTelescopeData. This section describes its data structure and usage.
Please assume that one of the name of TTelescopeData object is “tel1”
# after some process
artemis [*] br tel1
art::TTelescopeData
Data Members
TVector3 fPos detected position (X, Y, Z)
int fXID X strip number
int fYID Y strip number
int fNE number of all SSDs
double fdE energy at first layor
double fdEX X side energy (=~ fdEY)
double fdEY Y side energy (=~ fdEX)
double fE added energy at thick SSDs
double fEtotal all energy deposit in the telescope
double fTiming timing information at the first layor (X side)
double fYTiming for case that X side have trouble (Y side)
double fTheta_L reaction angle in LAB system
vector<double> fEnergyArray energy array for each SSD
vector<double> fTimingArray timing array for each SSD
ESortType kID
ESortType kTiming
ESortOrder kASC
ESortOrder kDESC
# snip for Method as for nowThese are the all data members of the “TTelescopeData”. The most commonly used variables are “fXID”, “fYID”, “fdE” and “fE”. Other variables are accessed by using methods (explain later). The meaning of these variables are written the upper code block.
We use them like,
artemis [*] tree->Draw("tel1.fYID:tel1.fXID>>strip(16,-0.5,15.5, 16,-0.5,15.5)","","colz")
artemis [*] tree->Draw("tel1.fdE:tel1.fE","","")or we can use in histogram definition file of course.
The following are the methods of the TTelescopeData object:
# after some process
artemis [*] br tel1
# snip for Data Members
Methods
TTelescopeData& operator=
TVector3 GetPosition
Double_t X
Double_t Y
Double_t Z
void SetPosition
void SetPosition
Int_t GetN
void SetN
Int_t GetXID
void SetXID
Int_t GetYID
void SetYID
Double_t GetdE
void SetdE
Double_t GetdEX
void SetdEX
Double_t GetdEY
void SetdEY
Double_t GetE
void SetE
Double_t GetEtotal
void SetEtotal
Double_t GetTelTiming
void SetTelTiming
Double_t GetTelYTiming
void SetTelYTiming
Double_t GetTheta_L
void SetTheta_L
Double_t A
DoubleVec_t GetEnergyArray
Double_t GetEnergyArray
void PushEnergyArray
DoubleVec_t GetTimingArray
Double_t GetTimingArray
void PushTimingArray
Double_t E
Double_t T
void Copy
void Clear
Bool_t CheckTObjectHashConsistency
See also
art::TDataObject base class for data objectThe most commonly used methods are “X()”, “Y()”, “Z()”, “E()”, “T()” and “A()”. There are also longer name methods, but it is troublesome to write long methods, so I prepared short name methods. The longer name methods are mainly used in the source processor to make it more readable.
X(): return fPos.X(), detected X positionY(): return fPos.Y(), detected Y positionZ(): return fPos.Z(), detected Z positionE(): return fEtotal, total energy deposit in the telescopeE(id: int): return fEnergyArray[id], energy deposit of each Si layer, id=0 means dE, id=1 means second layerT(): return fTiming, detected timing at first layerT(id: int): return fTimingArray[id], timing at the “id” th Si detectorA(): return fTheta_L, the angle of the event, deg unitWe use them like:
artemis [*] tree->Draw("tel1.Y():tel1.X()","","colz")
artemis [*] tree->Draw("tel1.E(0):tel1.E()","","colz")
artemis [*] tree->Draw("tel1.E():tel1.A()","","colz")In the physics run (production run or physics measurement), we check all the detector data like tracking detector (PPAC, MWDC), TOF (RF) and Telescope data. In order to analyse them, we prepared steering/chkf3.yaml steering file. This file includes all the information of F3 analysis.
$ artlogin (username)
$ a
artemis [0] add steering/chkf3.yaml NAME=hoge NUM=0000
artemis [1] res
artemis [2] sus
artemis [3] ls
artemis
> 0 art::TTreeProjGroup f3check f3check
1 art::TAnalysisInfo analysisInfo
2 art::TTreeProjGroup mwdca mwdca
3 art::TTreeProjGroup mwdcb mwdcb
4 art::TTreeProjGroup tel1 tel1
5 art::TTreeProjGroup tel2 tel2
6 art::TTreeProjGroup tel3 tel3
7 art::TTreeProjGroup tel4 tel4
8 art::TTreeProjGroup tel5 tel5
9 art::TTreeProjGroup tel6 tel6
10 art::TTreeProjGroup MUX MUX
11 TDirectory MWDCCalibHists MWDC calibration
# this is the situation at 2024/01/22There are many histogram definition, and you can freely update the histogram files.
Because this file contains most of the knowledge about the steering file, so you can test your own histogram or something based on this file.
$ cd steering
$ cp chkf3.yaml chkYourOwnName.yaml
$ vi (emacs) chkYourOwnName.yaml
# change or test something
$ acd
$ a
artemis [0] add steering/chkYourOwnName.yaml NAME=hoge NUM=0000
artemis [1] res
artmeis [2] sus
# some analysisIt is very important to select events for online analysis as well. There are several options to do so, and I will cover all of them. If you know other useful way, please let me know.
For clearer understanding, we will use this figure, and we call this as default figure:

We explained in the Histograms page, but again we describe about histogram definition focus on the cut part.
The default figure defined like this:
anchor:
- &f2ppacx ["f2ppac.fX",200,-50.,50.]
- &f2ppacy ["f2ppac.fY",200,-50.,50.]
alias:
group:
- name: f2check
title: f2_check
contents:
- name: f2PPAC X vs Y
title: f2ppac X vs Y
x: *f2ppacx
y: *f2ppacyFor example, let’s add the gate to select only “-10.0 < X < 10.0”. We can use alias node to define that.
anchor:
- &f2ppacx ["f2ppac.fX",200,-50.,50.]
- &f2ppacy ["f2ppac.fY",200,-50.,50.]
alias:
centerx: abs(f2ppac.fX) < 10.0;
group:
- name: f2check
title: f2_check
contents:
- name: f2PPAC X vs Y{centerx}
title: f2ppac X vs Y{centerx}
x: *f2ppacx
y: *f2ppacy
cut: centerxThen the following histogram is created.

Also, multiple conditions can be specified at the same time.
anchor:
- &f2ppacx ["f2ppac.fX",200,-50.,50.]
- &f2ppacy ["f2ppac.fY",200,-50.,50.]
alias:
centerx: abs(f2ppac.fX) < 10.0;
centery: abs(f2ppac.fY) < 10.0;
group:
- name: f2check
title: f2_check
contents:
- name: f2PPAC X vs Y{centerx && centery}
title: f2ppac X vs Y{centerx && centery}
x: *f2ppacx
y: *f2ppacy
cut: centerx && centery
We can also use TCutG object to select the event.
As in new commands page, let’s assume we create this TCutG ROOT file by tcutg command.
And the ROOT file and TCutG object name is f2star.

If you want to use this object to select event, just add this line is fine, as long as you use userlogon.C.
anchor:
- &f2ppacx ["f2ppac.fX",200,-50.,50.]
- &f2ppacy ["f2ppac.fY",200,-50.,50.]
alias:
group:
- name: f2check
title: f2_check
contents:
- name: f2PPAC X vs Y{f2star}
title: f2ppac X vs Y{f2star}
x: *f2ppacx
y: *f2ppacy
cut: f2star
Of course we can use in “tree->Draw()” because this TCutG objects are automatically loaded. The following sentence generate the same figure.
artemis [] fcd 0
artemis [] zo
artemis [] tree->Draw("f2ppac.fY:f2ppac.fX>>(200,-50.,50., 200,-50.,50.)","f2star","colz")The method used until now was to process just histograms and analyze them for all events in the event loop. If you know which events you don’t want, there are processors that allow you to skip the event loop under certain conditions. This may speed up the event loop.
For example. when we want to analyze only beam single event (it means the events are not coincident with SSD, and let’s suppose that condition is given by single.fID==0), let’s prepare the steering file.
Processor:
- name: proc_gateinit
type: art::TGateArrayInitializer
parameter:
OutputTransparency: 1
- name: proc_gate
type: art::TTreeFormulaGateProcessor
parameter:
Definitions:
- "beam_single; single.fID==0"
OutputTransparency: 1
Verbose: 1
- name: beam_single_gate
type: art::TGateStopProcessor
parameter:
GateName: beam_single
OutputTransparency: 1
StopIf: 0
Verbose: 1To use TGateStopProcessor, we need to initialize the “gate” array object, so the first art::TGateArrayInitializer is needed.
In the second processor, art::TTreeFormulaGateProcessor, we define the gate condition.
Then the art::TGateStopProcessor judges the event is skipped or not.
In the case of StopIf: 0, artemis ignore the event that the condition become false.
In other words, StopIf: 0 means artemis will analyze the event only when the condition is true.
Then, including this yaml file to the main steering file, you can check only the selected events.
Anchor:
- &input ridf/@NAME@@NUM@.ridf
- &output output/@NAME@/@NUM@/hoge@NAME@@NUM@.root
- &histout output/@NAME@/@NUM@/hoge@NAME@@NUM@.hist.root
Processor:
- name: timer
type: art::TTimerProcessor
- name: ridf
type: art::TRIDFEventStore
parameter:
OutputTransparency: 1
InputFiles:
- *input
SHMID: 0
- name: mapper
type: art::TMappingProcessor
parameter:
OutputTransparency: 1
# include some other steering files
- include: gate/coin.yaml
- name: outputtree
type: art::TOutputTreeProcessor
parameter:
FileName:
- *outputWe didn’t check if we can use the TCutG object in this process…
Artemis produces mainly TArtTree and the branches are TClonesArray(art::hoge).
It means that all objects rely on the artemis library, and we cannot open and check the data by using normal ROOT.
Also, sometimes it is necessary top check the raw data obtained by ADC and TDC as it is. Of course, the real raw data is binary and therefore difficult to read, so we will check the raw data after the decoders.
Related processors;
We already prepared conf/map files, but in this case, we need to prepare conf/seg files.
There are two files.
This is an example, so please change it according to the experimental conditions.
#modulename:
# id: module id (it is module-specific)
# ch: channel number
# values:
# - name1: [Nbin: int, min: double, max: double] it is for 2D histogram (these values vs. ch)
# - name2: [Nbin: int, min: double, max: double] <= somehow two line needed...?
MADC32:
id: 32
ch: 32
values:
- adc: [4000, 0., 4000.]
- tdc: [4000, 0., 4000.] # no use, but seems it needed...
V1190A:
id: 24
ch: 128
values:
- tdcL: [300, -5000., 300000.]
- tdcT: [300, -5000., 300000.]The module id list is here.
#segment_name:
# segid: [[dev], [fp], [det]] <= same as a map file
# type: V7XX <= defined type in modulelist.yaml
# modules:
# - id: geo1
# - id: geo2
V1190:
segid: [12, 0, 7]
type: V1190A
modules:
- id: 0
- id: 1
MADC:
segid: [12, 1, 6]
type: MADC32
modules:
- id: 0
- id: 1
- id: 2Based on these two conf file, the steering/chkseg.yaml file produce rawdata histograms and TTree object.
We can use steering/chkseg.yaml without any change I think.
This is an example from one CRIB experiment.
$ a
artemis [0] add steering/chkseg.yaml NAME=hoge NUM=0000
artemis [1] res
artemis [2] sus
artemis [3] ls
artemis
> 0 TDirectory SegmentHistogram art::TSegmentCheckProcessor
artemis [4] cd 0
artemis [5] ls
SegmentHistogram
> 0 TDirectory E7_V1190 E7_V1190
1 TDirectory J1_V785 J1_V785
2 TDirectory J1_MADC J1_MADC
3 TDirectory J1_V1190 J1_V1190
artemis [6] cd 0
artemis [7] ls
E7_V1190
> 0 TH2F E7_V1190_0_tdcL E7_V1190_0_tdcL
1 TH2F E7_V1190_0_tdcT E7_V1190_0_tdcT
2 TH2F E7_V1190_1_tdcL E7_V1190_1_tdcL
3 TH2F E7_V1190_1_tdcT E7_V1190_1_tdcT
# we can check these histograms by ht command
artemis [8] fcd 0
artemis [9] br
E7_V1190_0 vector<vector<int> >
E7_V1190_1 vector<vector<int> >
J1_V785_0 vector<int>
J1_V785_1 vector<int>
J1_V785_2 vector<int>
J1_MADC_3 vector<int>
J1_MADC_4 vector<int>
J1_MADC_5 vector<int>
J1_V1190_0 vector<vector<int> >If the module ID = 24 or 25, it is multihit TDC, so the branch become 2D vector. When you want to use std::vector method, you can use by “@” like
artemis [*] tree->Draw("J1_V785_0@.size()")Of course we can open this output ROOT file from normal ROOT.
This section explain the example of the offline analysis (some useful processors).
A brief example of an analysis flow using artemis_crib is given below. This is just the method recommended by okawak, so you can proceed with the analysis as you like.
I hope this page will give you some ideas.
The acquired data contains a lot of noise and other data that is not absolutely necessary for analysis,
and if you use artemis to do an event loop for all events, it will take a long time.
Therefore, a root file should only be created for a specific RUN (data file) to save time.
(It is tempting to look at the data using with a lot of statistics,
but we believe that this is done by online-analysis and concentrate on reducing useless data.)
$ artlogin (user)
$ a
artemis [] a steering/hoge.yaml NAME=run NUM=0000
artemis [] .qAs you want to look at a variety of data I think, it is convenient to create a canvas each time with ‘tree->Draw()’, instead of defining a histogram. Not defining a histogram also has the advantage that the event loop is faster.
Once the root file has been created, it can be read directly from that file, eliminating the waiting time.
$ a output/created_rootfile_path.root
artemis [] tree->Draw("hoge")It is good idea to make a “.C” macro file to record and reproduce the analysis process.
Here are some points I think would be good to check:
If there are problems during the experiment, there may be many other things to check.
It is possible that some parameter is different for each RUN. There are many possibilities, for example, a trouble may have occurred and the conditions of the detector have changed. In such cases, it is necessary to take action, for example, to prepare several calibration parameters.
While some of these changes in parameters can be predicted in advance, unforeseen changes can also occur.
These are the problems I have encountered, but others with different tendencies may exist.
If you can anticipate it, it is good to have a plan for dealing with it at this point and move on to the next step. However, there is no need to waste time here as it can be noticed after merging the overall measurements in the following step.
This section explain the example of the Monte Carlo simulation by using artemis.
It is very useful to use Monte Carlo (MC) simulation to obtain various calculated values. From this section, we will show the simple example how to perform MC simulation using artemis.
First, we will set the generator of the beam that will bombard on the target. We have two kinds of generator:
After you get beam information from a measurement, second option might be useful.
This is an example of a steering file.
Processor:
- name: random_eventstore
type: art::TRandomNumberEventStore
parameter:
OutputTransparency: 1
MaxLoop: *loopnum
- name: beam_generator
type: art::TRandomBeamGenerator
parameter:
OutputCollection: *beam_name
OutputTrackCollection: *track_name
#beam particle information
MassNum: *beam_A
AtomicNum: *beam_Z
ChargeNum: *beam_Z
IniEnergy: *beam_E
#beam tracking information
Xsigma: 1.0 # mm
Ysigma: 1.0 # mm
Asigma: 1.0 # deg
Bsigma: 1.0 # deg
Esigma: *beam_EsigmaThis produces two branch, “beam” and “track”.
Actually “track” information is contained in “beam” branch.
Here is the detail of the type of “beam”.
artemis [] classinfo art::TParticleInfo
art::TParticleInfo
Data Members
int fMassNumber
int fAtomicNumber
int fCharge
double fEnergy kinetic energy in LAB system
double fCurrentZ current Z position
double fTime Duration time (ns)
TTrack fTrack tracking information in LAB system
TLorentzVector fVec lorentz vector (px, py, pz, E) of this particle
double fTheta_cm theta angle (deg) in CM system
double fPhi_cm phi angle (deg) in CM system
-- snip --fCurrentZ is the current Z position and at the beam generation, this value will be set as 0 after this processor.
For example, the beam angle distribution will be like this.
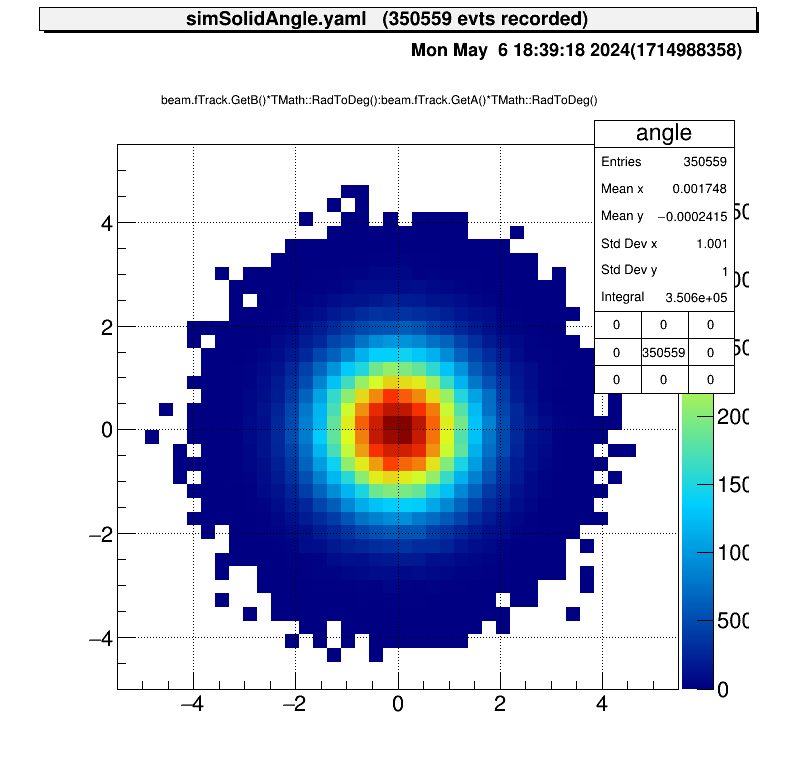
The tracking information obtained from the experiment can also be used to generate the beam. In the current version, the position and angle of the beam are read from a ROOT file and the energy is given randomly.
This is an example of a steering file.
Processor:
- name: periodic_tree
type: art::TTreePeriodicEventStore
parameter:
OutputTransparency: 1
FileName: *input
TreeName: tree
MaxEventNum: *loopnum
- name: proc_copy_processor
type: art::TBranchCopyProcessor
parameter:
InputCollection: f3ppac # need to inherit from TTrack
OutputCollection: *track_name
- name: beam_generator
type: art::TTreeBeamGenerator
parameter:
InputCollection: *track_name
OutputCollection: *beam_name
# beam particle information
MassNum: *beam_A
AtomicNum: *beam_Z
ChargeNum: *beam_Z
IniEnergy: *beam_E
Esigma: *beam_EsigmaIt reads an object named “f3ppac”, which inherits from TTrack, from a TTree named “tree” in a file named “*input”,
and creates an object of the same type as before named TParticleInfo.
make a reaction considering phase space.
- name: reaction_proc
type: art::TNBodyReactionProcessor
parameter:
InputCollection: *beam_name
OutputCollection: products # size is DecayParticleNum
OutputReactionCollection: reaction
# beam information (for initialize TSrim)
BeamNucleus: [*beam_Z, *beam_A] # (Z, A)
BeamEnergy: *beam_E
# target information
TargetIsGas: true # false-> solid, true-> gas target
TargetName: he # from TSrim energy loss
TargetMassNum: 4 # hit ion
TargetAtomicNum: 2 # hit ion
TargetThickness: 1000 # mm (for gas target, allow up to this value)
TargetPressure: 250 # Torr (used for gas target)
# reaction particles information
DecayParticleNum: 2
ReactionMassNum: [29, 1] # will be [id=0, id=1]
ReactionAtomicNum: [15, 1]
ExciteLevel: [0.0, 0.0] # MeV
# cross section file, if not, it use constant cross section for energy
# require: "energy cross-section" format, deliminator should be a space ' '
CrossSectionPath: path/to/cs/file
CrossSectionType: 0 # 0-> LAB, kinematics is different, 1-> LAB, kinematics is same, 2-> CMNOTE
TargetName that registered this library# comment
# energy cross_section
0.0 0.0
0.1 1.0 # comment
# 0.2, 2.0 camma is not allowed...products is size two array, like products[0] -> id=0 particlereaction contain reaction information, like Ecm, Thetacmartemis [] classinfo art::TReactionInfo
art::TReactionInfo
Data Members
double fEnergy / Ecm of the reaction
double fTheta / Thetacm of the reaction
double fX / reaction position at LAB system, x
double fY / reaction position at LAB system, y
double fZ / reaction position at LAB system, z
double fExEnergy / excited energy of residual nucleus
ESortType kID
ESortType kTiming
ESortOrder kASC
ESortOrder kDESC We will show an example of a simulation performed for a gas target case like the steering file above. (of course we can use it also for solid target case!)
Details of the reactions that took place are shown in these figures.
The reaction cross section files are appropriately specified.
This figure is the reaction energy at CM system.
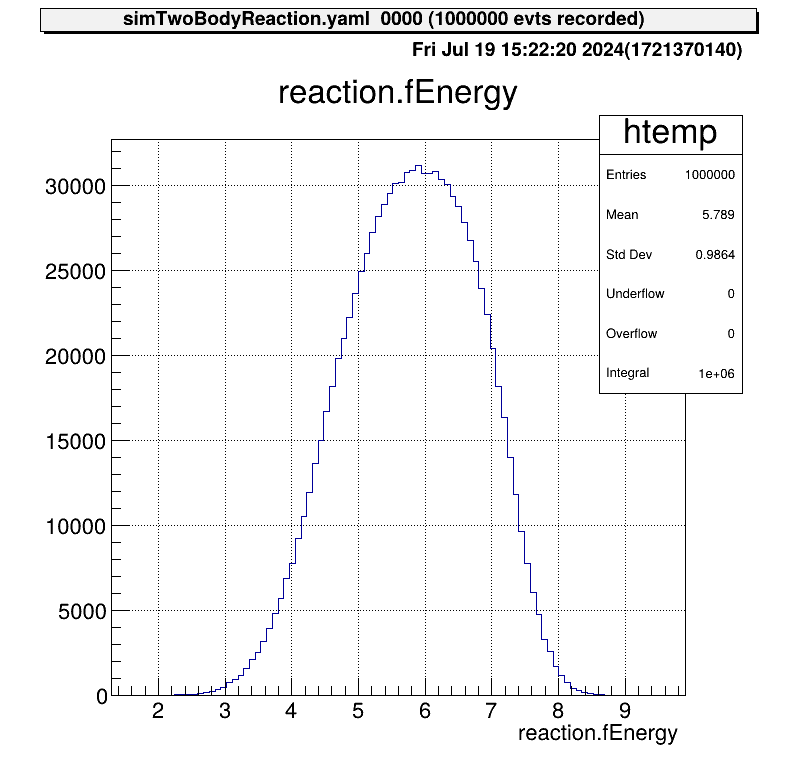
This distribution was created from a file of reaction cross section. The effect of beam energy spread is included, so the edge of the peak is not sharp.
According to this energy distribution, the distribution of the positions of the reactions is as follows.
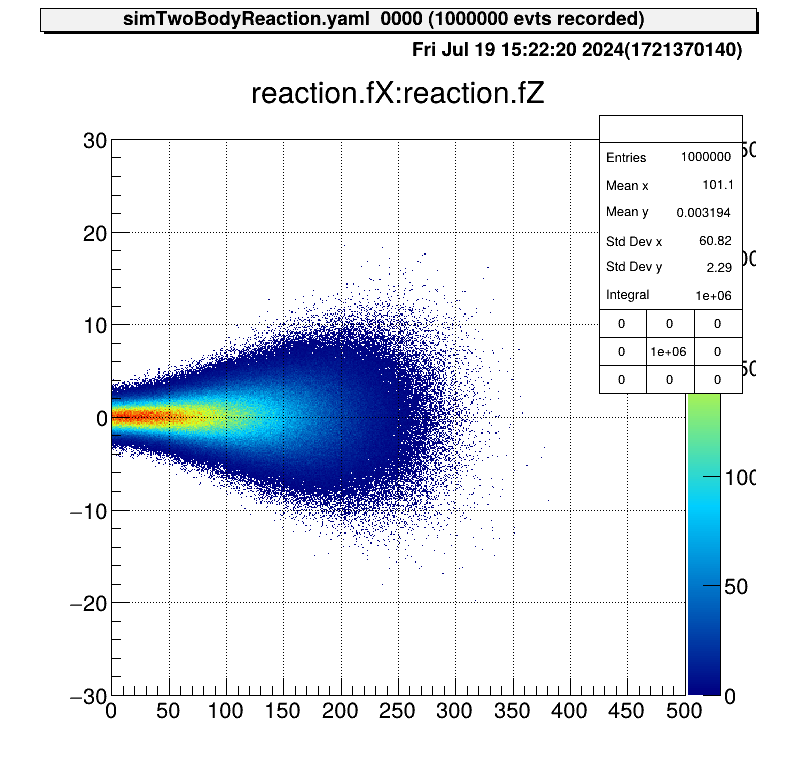
The angular distribution at CM system of reactions is assumed to be uniform. This figure shows the direction of one particle that is produced from this reaction.
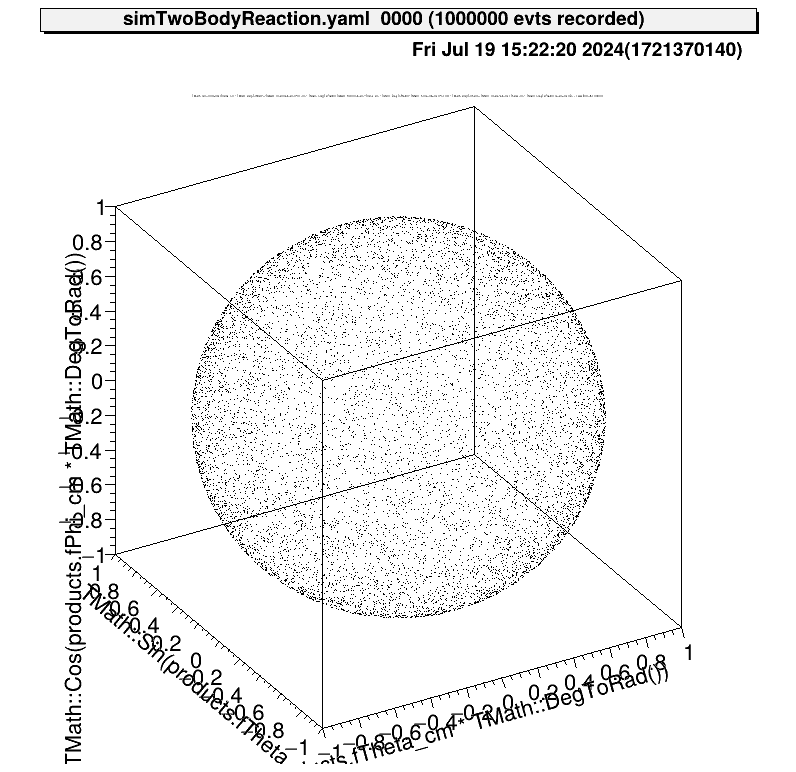
Also the relationship between the kinematics of the reactions will be like this.
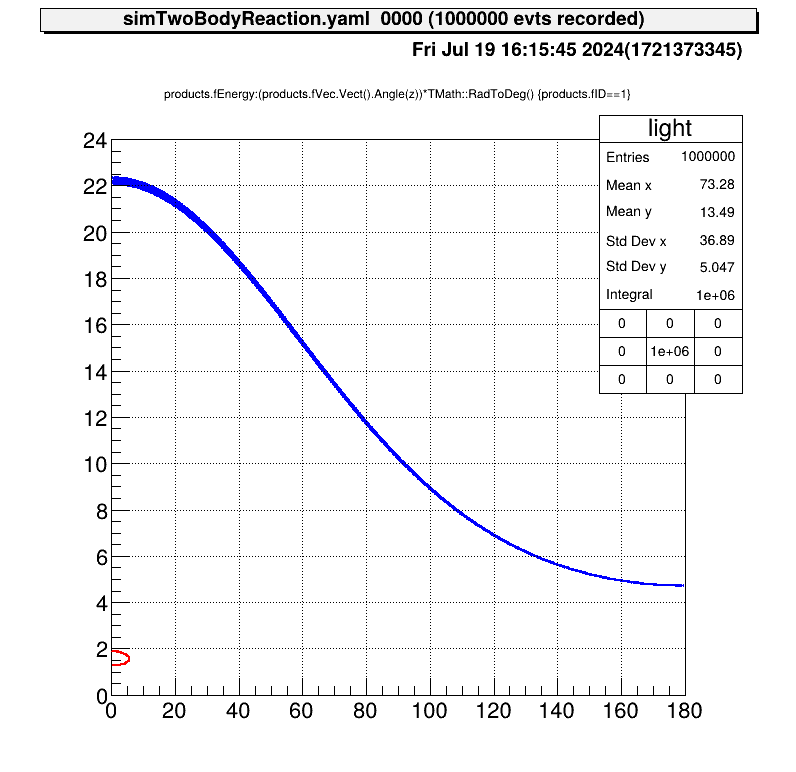
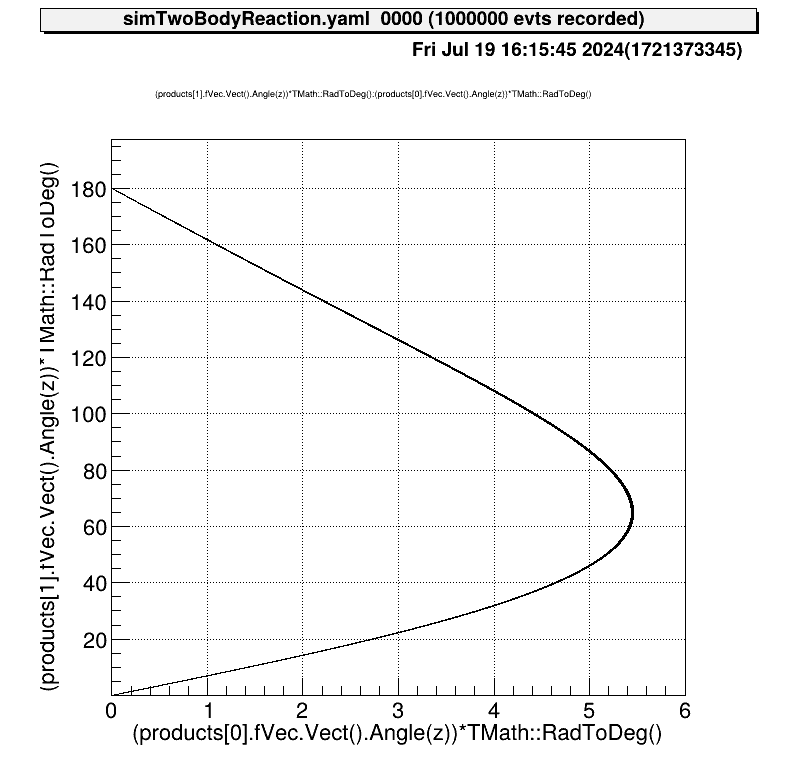
Configure detector geometry to judge if the reaction particle hit the detector or not.
material:
- name: Vaccum # id=0
atomic_mass: 0.0
atomic_num: 0.0
density: 0.0 # g/cm3
- name: Si # id=1
atomic_mass: 28.084
atomic_num: 14.0
density: 2.321
# Note: beam axis -> z, upper direction -> y
conposition:
detector:
- name: tel1
strip: [16, 16]
center_rotation: [0., 0., 322.0] # mm
offset: [0., 0., 0.]
distance: 244.0
angle: -4.0 # deg
thickness: [0.02, 0.301, 1.494, 1.486]
pedestal: [0., 0., 0., 0.] # MeV, below this energy, assume 0 MeV
material: [Si]
- name: tel2
strip: [16, 16]
center_rotation: [0., 0., 322.0]
offset: [0., 0., 0.]
distance: 154.5
angle: 27.0
thickness: [0.02, 0.300, 1.494, 1.485]
pedestal: [0., 0., 0., 0.] # MeV, below this energy, assume 0 MeV
material: [Si]
volume:
top: # detector world
name: TOP
type: box # now only box is available
material: 0
size: [400.0, 200.0, 1288.0] # mm
detector:
- name: tel1
type: box
material: 1
size: [50.0, 50.0, 1.0] # mm
- name: tel2
type: box
material: 1
size: [50.0, 50.0, 1.0] # mmThe geometry information is used only for the judgement, so any material is actually okay.
The material node is used to define TGeoMaterial and TGeoMedium classes.
From name to density node are used to make a instance of this object.
This values are not used in the current processors.
The next conposition node define the detector configuration.
General telescopes of the CRIB experiment consist of DSSSD and SSD (single-pad),
and the node below defines the SSD of the telescope.
SRIMlib calculation. This node is defined as a array for each layer, but if it is one, the same material is applied. For example, in example.yaml, [Si] means [Si, Si, Si, Si]. (You need to prepare SRIMlib setting beforehand!)As for the node is center_rotation, offset, distance and angle, please see this figure.

Please note that the center_rotation and offset are defined in (x, y, z) coordinate, and distance and angle are scalar value.
The unit of length is mm, angle is deg.
The sign of the angle is defined as positive at this figure (right-hand system). And generally, we set z=0 at target position. (For the gas target, we set 0 at window position.)
The last part volume node, the shape of the detector will be defined by using TGeoVolume class.
The TGeoVolume needs name, type, material and size.
For the type, I only prepared “box”. (It means the code use only vol->MakeBox method.)
The first top node must be set because it defined “detector world”.
Generally, the material is okay to set vaccum.
And the material is defined in the material node, and the id (the order) can be used.
So the format is like material: 0.
And the size is generally set to the size of the scattering chamber, but for the safety, it is okay to set larger number.
Also the unit is mm and format is (x, y, z).
Next, at the volume/detector node, we can define the detector size.
NOTE: the volume/detector/name should be the same with conposition/detector/name node.
Then, let’s check if the parameter file can be correctly used.
Anchor:
Processor:
- name: detector_initialize
type: art::TUserGeoInitializer
parameter:
FileName: prm/geo/example.yaml
Visible: true
OutputTransparency: 1This steering file doesn’t use event loop. Just we want to check the parameter file works well or not.
Then let’s see in the artemis!
$ acd
$ a
-- snip --
artemis [0] add steering/geo_example.yaml
-- snip --
artemis [1] ls
artemis
> 0 TGeoVolume TOP Top volume
artemis [2]The detector geometry object is successfully generated! In order to check the object, please use draw command for example. (It is defined only in CRIB artemis, to draw not only histogram object. This is under development.)
Or you can get the object with some methods and obj->Draw() will work.
artemis [2] draw 0
The red box is the TOP, and the black boxes are detectors. If the detector is placed where you expect it to be, the parameters have been successfully set!
Using the defined reactions (TNBodyReactionProcessor) and geometry (TUserGeoInitialiser) so far, it is possible to determine whether the particles enter the detector after the reaction and to simulate the parameters obtained.
- name: detector_proc
type: art::TDetectParticleProcessor
parameter:
InputCollection: products
InputTrackCollection: track
OutputCollection: detects
# target information (use if gas target)
TargetIsGas: *target_is_gas # true -> gas target
TargetName: *target_name
TargetPressure: *target_pressure # Torr (used for gas target)
EnergyResolution: [0.0] # x 100 = %, det id = 0, 1, ..products is an array object of produced ions [id=0 ion, id=1 ion…]detects is an array object of detected ions [id=0 ion, id=1 ion…]We will show an example of a simulation performed for a gas target case, and resolution = 0.0.
First, this is a figure of the detected position of light particle. We defined five telescopes in this example, and we can clearly see the position of the detectors. If we focus on heavy particle, we cannot see the position distribution because all ions are stopped in the gas target.

As an example, we will show you figures of a telescope with the largest angle. Here is dE-E plots.

The relationship between energy and timing is like this.

Also, here is the angular distribution of the LAB system.

The processor created by okawak allows simple simulations to be performed without creating new sources, if the following conditions are met.
There are only two files that need to be prepared for the simulation.
prm/geo/hoge.yaml -> define detector configuration (example prm/geo/si26a.yaml)
currentsteering/huga.yaml -> define simulation parameters (example steering/simNBodyReaction.yaml)As an application of the above four sections, I would like to explain how to calculate solid angles using Monte Carlo methods!